Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations
- PMID: 23593013
- PMCID: PMC3617151
- DOI: 10.1371/journal.pgen.1003390
Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations
Abstract
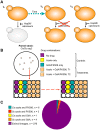
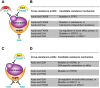
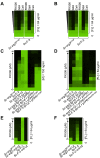
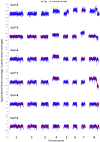
The evolution of drug resistance in fungal pathogens compromises the efficacy of the limited number of antifungal drugs. Drug combinations have emerged as a powerful strategy to enhance antifungal efficacy and abrogate drug resistance, but the impact on the evolution of drug resistance remains largely unexplored. Targeting the molecular chaperone Hsp90 or its downstream effector, the protein phosphatase calcineurin, abrogates resistance to the most widely deployed antifungals, the azoles, which inhibit ergosterol biosynthesis. Here, we evolved experimental populations of the model yeast Saccharomyces cerevisiae and the leading human fungal pathogen Candida albicans with azole and an inhibitor of Hsp90, geldanamycin, or calcineurin, FK506. To recapitulate a clinical context where Hsp90 or calcineurin inhibitors could be utilized in combination with azoles to render resistant pathogens responsive to treatment, the evolution experiment was initiated with strains that are resistant to azoles in a manner that depends on Hsp90 and calcineurin. Of the 290 lineages initiated, most went extinct, yet 14 evolved resistance to the drug combination. Drug target mutations that conferred resistance to geldanamycin or FK506 were identified and validated in five evolved lineages. Whole-genome sequencing identified mutations in a gene encoding a transcriptional activator of drug efflux pumps, PDR1, and a gene encoding a transcriptional repressor of ergosterol biosynthesis genes, MOT3, that transformed azole resistance of two lineages from dependent on calcineurin to independent of this regulator. Resistance also arose by mutation that truncated the catalytic subunit of calcineurin, and by mutation in LCB1, encoding a sphingolipid biosynthetic enzyme. Genome analysis revealed extensive aneuploidy in four of the C. albicans lineages. Thus, we identify molecular determinants of the transition of azole resistance from calcineurin dependence to independence and establish multiple mechanisms by which resistance to drug combinations evolves, providing a foundation for predicting and preventing the evolution of drug resistance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90.PLoS Pathog. 2010 Aug 26;6(8):e1001069. doi: 10.1371/journal.ppat.1001069. PLoS Pathog. 2010. PMID: 20865172 Free PMC article.
-
Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin.PLoS Pathog. 2009 Jul;5(7):e1000532. doi: 10.1371/journal.ppat.1000532. Epub 2009 Jul 31. PLoS Pathog. 2009. PMID: 19649312 Free PMC article.
-
Metabolic control of antifungal drug resistance.Fungal Genet Biol. 2010 Feb;47(2):81-93. doi: 10.1016/j.fgb.2009.07.004. Epub 2009 Jul 10. Fungal Genet Biol. 2010. PMID: 19595784
-
Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach.Virulence. 2017 Feb 17;8(2):186-197. doi: 10.1080/21505594.2016.1201250. Epub 2016 Jun 20. Virulence. 2017. PMID: 27325145 Free PMC article. Review.
-
Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans.J Antimicrob Chemother. 2020 Feb 1;75(2):257-270. doi: 10.1093/jac/dkz400. J Antimicrob Chemother. 2020. PMID: 31603213 Free PMC article. Review.
Cited by
-
Synergy Maps: exploring compound combinations using network-based visualization.J Cheminform. 2015 Aug 1;7:36. doi: 10.1186/s13321-015-0090-6. eCollection 2015. J Cheminform. 2015. PMID: 26236402 Free PMC article.
-
Targeting fungal membrane homeostasis with imidazopyrazoindoles impairs azole resistance and biofilm formation.Nat Commun. 2022 Jun 25;13(1):3634. doi: 10.1038/s41467-022-31308-1. Nat Commun. 2022. PMID: 35752611 Free PMC article.
-
Ploidy-regulated variation in biofilm-related phenotypes in natural isolates of Saccharomyces cerevisiae.G3 (Bethesda). 2014 Jul 24;4(9):1773-86. doi: 10.1534/g3.114.013250. G3 (Bethesda). 2014. PMID: 25060625 Free PMC article.
-
Penetrating cations induce pleiotropic drug resistance in yeast.Sci Rep. 2018 May 25;8(1):8131. doi: 10.1038/s41598-018-26435-z. Sci Rep. 2018. PMID: 29802261 Free PMC article.
-
Molecular Characterization of Gβ-Like Protein CpcB Involved in Antifungal Drug Susceptibility and Virulence in A. fumigatus.Front Microbiol. 2016 Feb 9;7:106. doi: 10.3389/fmicb.2016.00106. eCollection 2016. Front Microbiol. 2016. PMID: 26903985 Free PMC article.
References
-
- Chopra I (2012) The 2012 Garrod Lecture: Discovery of antibacterial drugs in the 21st century. J Antimicrob Chemother 68 3: 496–505 doi:10.1093/jac/dks436. - DOI - PubMed
-
- Palumbi SR (2001) Humans as the world's greatest evolutionary force. Science 293: 1786–1790. - PubMed
-
- Cowen LE (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol 6: 187–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

