An essential role for zygotic expression in the pre-cellular Drosophila embryo
- PMID: 23593026
- PMCID: PMC3616919
- DOI: 10.1371/journal.pgen.1003428
An essential role for zygotic expression in the pre-cellular Drosophila embryo
Abstract
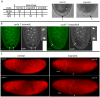
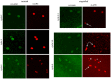
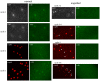
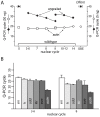
The Drosophila embryo proceeds through thirteen mitotic divisions as a syncytium. Its nuclei distribute in the embryo's interior during the first six divisions, dividing synchronously with a cycle time of less than ten minutes. After seven divisions (nuclear cycle 8), the syncytial blastoderm forms as the nuclei approach the embryo surface and slow their cycle time; subsequent divisions proceed in waves that initiate at the poles. Because genetic studies have not identified zygotic mutants that affect the early divisions and because transcription has not been detected before cycle 8, the early, pre-blastoderm embryo has been considered to rely entirely on maternal contributions and to be transcriptionally silent. Our studies identified several abnormal phenotypes in live engrailed (en) mutant embryos prior to cycle 8, as well as a small group of genes that are transcribed in embryos prior to cycle 7. Nuclei in en embryos divide asynchronously, an abnormality that was detected as early as nuclear cycle 2-3. Anti-En antibody detected nuclear En protein in embryos at cycle 2, and expression of an En:GFP fusion protein encoded in the paternal genome was also detected in cycle 2 nuclei. These findings demonstrate that the Drosophila embryo is functionally competent for gene expression prior to the onset of its rapid nuclear divisions and that the embryo requires functions that are expressed in the zygote in order to faithfully prosecute its early, pre-cellularization mitotic cycles.
Conflict of interest statement
MBE is a cofounder and member of the Board of Directors of PLOS. The other authors have declared that no competing interests exist.
Figures



References
-
- Pritchard DK, Schubiger G (1996) Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev 10: 1131–1142. - PubMed
-
- ten Bosch JR, Benavides JA, Cline TW (2006) The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133: 1967–1977. - PubMed
-
- Anderson KV, Lengyel JA (1981) Changing rates of DNA and RNA synthesis in Drosophila embryos. Dev Biol 82: 127–138. - PubMed
-
- Edgar BA, Schubiger G (1986) Parameters controlling transcriptional activation during early Drosophila development. Cell 44: 871–877. - PubMed
-
- Zalokar M (1976) Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol 49: 425–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

