Effective identification of bacterial type III secretion signals using joint element features
- PMID: 23593149
- PMCID: PMC3617162
- DOI: 10.1371/journal.pone.0059754
Effective identification of bacterial type III secretion signals using joint element features
Abstract
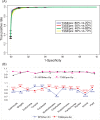
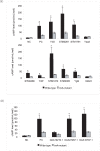
Type III secretion system (T3SS) plays important roles in bacteria and host cell interactions by specifically translocating type III effectors into the cytoplasm of the host cells. The N-terminal amino acid sequences of the bacterial type III effectors determine their specific secretion via type III secretion conduits. It is still unclear as to how the N-terminal sequences guide this specificity. In this work, the amino acid composition, secondary structure, and solvent accessibility in the N-termini of type III and non-type III secreted proteins were compared and contrasted. A high-efficacy mathematical model based on these joint features was developed to distinguish the type III proteins from the non-type III ones. The results indicate that secondary structure and solvent accessibility may make important contribution to the specific recognition of type III secretion signals. Analysis also showed that the joint feature of the N-terminal 6(th)-10(th) amino acids are especially important for guiding specific type III secretion. Furthermore, a genome-wide screening was performed to predict Salmonella type III secreted proteins, and 8 new candidates were experimentally validated. Interestingly, type III secretion signals were also predicted in gram-positive bacteria and yeasts. Experimental validation showed that two candidates from yeast can indeed be secreted through Salmonella type III secretion conduit. This research provides the first line of direct evidence that secondary structure and solvent accessibility contain important features for guiding specific type III secretion. The new software based on these joint features ensures a high accuracy (general cross-validation sensitivity of ∼96% at a specificity of ∼98%) in silico identification of new type III secreted proteins, which may facilitate our understanding about the specificity of type III secretion and the evolution of type III secreted proteins.
Conflict of interest statement
Figures




References
-
- Lindeberg M, Collmer A (2009) Gene ontology for type III effectors: capturing processes at the host-pathogen interface. Trend. Microbiol. 17: 304–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

