Adaptation of mycobacteria to growth conditions: a theoretical analysis of changes in gene expression revealed by microarrays
- PMID: 23593152
- PMCID: PMC3625197
- DOI: 10.1371/journal.pone.0059883
Adaptation of mycobacteria to growth conditions: a theoretical analysis of changes in gene expression revealed by microarrays
Abstract
Background: Microarray analysis is a powerful technique for investigating changes in gene expression. Currently, results (r-values) are interpreted empirically as either unchanged or up- or down-regulated. We now present a mathematical framework, which relates r-values to the macromolecular properties of population-average cells. The theory is illustrated by the analysis of published data for two species; namely, Mycobacterium bovis BCG Pasteur and Mycobacterium smegmatis mc(2) 155. Each species was grown in a chemostat at two different growth rates. Application of the theory reveals the growth rate dependent changes in the mycobacterial proteomes.
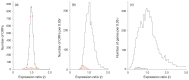
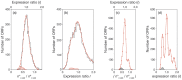
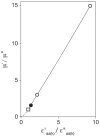
Principal findings: The r-value r (i) of any ORF (ORF(i)) encoding protein p (i) was shown to be equal to the ratio of the concentrations of p (i) and so directly proportional to the ratio of the numbers of copies of p (i) per population-average cells of the two cultures. The proportionality constant can be obtained from the ratios DNA: RNA: protein. Several subgroups of ORFs were identified because they shared a particular r-value. Histograms of the number of ORFs versus the expression ratio were simulated by combining the particular r-values of several subgroups of ORFs. The largest subgroup was ORF(j) (r (j) = 1.00± SD) which was estimated to comprise respectively 59% and 49% of ORFs of M. bovis BCG Pasteur and M. smegmatis mc(2) 155. The standard deviations reflect the properties of the cDNA preparations investigated.
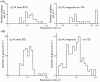
Significance: The analysis provided a quantitative view of growth rate dependent changes in the proteomes of the mycobacteria studied. The majority of the ORFs were found to be constitutively expressed. In contrast, the protein compositions of the outer permeability barriers and cytoplasmic membranes were found to be dependent on growth rate; thus illustrating the response of bacteria to their environment. The theoretical approach applies to any cultivatable bacterium under a wide range of growth conditions.
Conflict of interest statement
Figures
References
-
- Kendall SL, Rison SC, Movahedzadeh F, Frita R, Stoker NG (2004a) What do microarrays really tell us about Mycobacterium tuberculosis?. Trends Microbiol 12: 537–544. - PubMed
-
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474. - PubMed
-
- Southern EM (2001) DNA microarrays. History and overview Methods Mol Biol. 170: 1–15. - PubMed
-
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. - PubMed
-
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, et al. (2001) Massive gene decay in the leprosy bacillus. Nature 409: 1007–1011. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

