MrBayes tgMC³: a tight GPU implementation of MrBayes
- PMID: 23593277
- PMCID: PMC3621901
- DOI: 10.1371/journal.pone.0060667
MrBayes tgMC³: a tight GPU implementation of MrBayes
Abstract
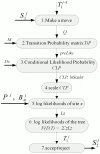
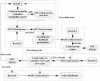
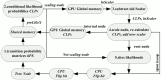
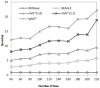
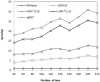
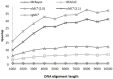
MrBayes is model-based phylogenetic inference tool using Bayesian statistics. However, model-based assessment of phylogenetic trees adds to the computational burden of tree-searching, and so poses significant computational challenges. Graphics Processing Units (GPUs) have been proposed as high performance, low cost acceleration platforms and several parallelized versions of the Metropolis Coupled Markov Chain Mote Carlo (MC(3)) algorithm in MrBayes have been presented that can run on GPUs. However, some bottlenecks decrease the efficiency of these implementations. To address these bottlenecks, we propose a tight GPU MC(3) (tgMC(3)) algorithm. tgMC(3) implements a different architecture from the one-to-one acceleration architecture employed in previously proposed methods. It merges multiply discrete GPU kernels according to the data dependency and hence decreases the number of kernels launched and the complexity of data transfer. We implemented tgMC(3) and made performance comparisons with an earlier proposed algorithm, nMC(3), and also with MrBayes MC(3) under serial and multiply concurrent CPU processes. All of the methods were benchmarked on the same computing node from DEGIMA. Experiments indicate that the tgMC(3) method outstrips nMC(3) (v1.0) with speedup factors from 2.1 to 2.7×. In addition, tgMC(3) outperforms the serial MrBayes MC(3) by a factor of 6 to 30× when using a single GTX480 card, whereas a speedup factor of around 51× can be achieved by using two GTX 480 cards on relatively long sequences. Moreover, tgMC(3) was compared with MrBayes accelerated by BEAGLE, and achieved speedup factors from 3.7 to 5.7×. The reported performance improvement of tgMC(3) is significant and appears to scale well with increasing dataset sizes. In addition, the strategy proposed in tgMC(3) could benefit the acceleration of other Bayesian-based phylogenetic analysis methods using GPUs.
Conflict of interest statement
Figures

Similar articles
-
MrBayes on a graphics processing unit.Bioinformatics. 2011 May 1;27(9):1255-61. doi: 10.1093/bioinformatics/btr140. Epub 2011 Mar 16. Bioinformatics. 2011. PMID: 21414986
-
MrBayes tgMC3++: A High Performance and Resource-Efficient GPU-Oriented Phylogenetic Analysis Method.IEEE/ACM Trans Comput Biol Bioinform. 2016 Sep-Oct;13(5):845-854. doi: 10.1109/TCBB.2015.2495202. Epub 2015 Oct 27. IEEE/ACM Trans Comput Biol Bioinform. 2016. PMID: 26529779
-
Efficient implementation of MrBayes on multi-GPU.Mol Biol Evol. 2013 Jun;30(6):1471-9. doi: 10.1093/molbev/mst043. Epub 2013 Mar 14. Mol Biol Evol. 2013. PMID: 23493260 Free PMC article.
-
GPU MrBayes V3.1: MrBayes on Graphics Processing Units for Protein Sequence Data.Mol Biol Evol. 2015 Sep;32(9):2496-7. doi: 10.1093/molbev/msv129. Epub 2015 May 26. Mol Biol Evol. 2015. PMID: 26012905
-
MGUPGMA: A Fast UPGMA Algorithm With Multiple Graphics Processing Units Using NCCL.Evol Bioinform Online. 2017 Oct 4;13:1176934317734220. doi: 10.1177/1176934317734220. eCollection 2017. Evol Bioinform Online. 2017. PMID: 29051701 Free PMC article. Review.
Cited by
-
Episodic evolution of a eukaryotic NADK repertoire of ancient provenance.PLoS One. 2019 Aug 1;14(8):e0220447. doi: 10.1371/journal.pone.0220447. eCollection 2019. PLoS One. 2019. PMID: 31369599 Free PMC article.
References
-
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. - PubMed
-
- Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20: 406–416.
-
- Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Mol Biol Evol 17: 368–376. - PubMed
-
- Hastings WK (1970) Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika 57(1): 97–109.
-
- Yang ZH, Rannala B (2012) Molecular phylogenetics: principles and practice. Nat Rev Genet 13: 303–314. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

