Klebsiella phage vB_KleM-RaK2 - a giant singleton virus of the family Myoviridae
- PMID: 23593293
- PMCID: PMC3622015
- DOI: 10.1371/journal.pone.0060717
Klebsiella phage vB_KleM-RaK2 - a giant singleton virus of the family Myoviridae
Erratum in
- PLoS One. 2013;8(12). doi:10.1371/annotation/a1d15675-2942-41ba-92f4-3dad6bc6cac6
Abstract
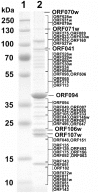
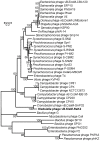
At 346 kbp in size, the genome of a jumbo bacteriophage vB_KleM-RaK2 (RaK2) is the largest Klebsiella infecting myovirus genome sequenced to date. In total, 272 out of 534 RaK2 ORFs lack detectable database homologues. Based on the similarity to biologically defined proteins and/or MS/MS analysis, 117 of RaK2 ORFs were given a functional annotation, including 28 RaK2 ORFs coding for structural proteins that have no reliable homologues to annotated structural proteins in other organisms. The electron micrographs revealed elaborate spike-like structures on the tail fibers of Rak2, suggesting that this phage is an atypical myovirus. While head and tail proteins of RaK2 are mostly myoviridae-related, the bioinformatics analysis indicate that tail fibers/spikes of this phage are formed from podovirus-like peptides predominantly. Overall, these results provide evidence that bacteriophage RaK2 differs profoundly from previously studied viruses of the Myoviridae family.
Conflict of interest statement
Figures



References
-
- Hendrix RW (2002) Bacteriophages: evolution of the majority. Theor Popul Biol 61: 471–480. - PubMed
-
- Ackerman H-W (2011) Bacteriophage taxonomy. Cambridge Publishing, Microbiology. Australia.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

