Nutrigenomics in Arma chinensis: transcriptome analysis of Arma chinensis fed on artificial diet and Chinese oak silk moth Antheraea pernyi pupae
- PMID: 23593338
- PMCID: PMC3623872
- DOI: 10.1371/journal.pone.0060881
Nutrigenomics in Arma chinensis: transcriptome analysis of Arma chinensis fed on artificial diet and Chinese oak silk moth Antheraea pernyi pupae
Abstract
Background: The insect predator, Arma chinensis, is capable of effectively controlling many pests, such as Colorado potato beetle, cotton bollworm, and mirid bugs. Our previous study demonstrated several life history parameters were diminished for A. chinensis reared on an artificial diet compared to a natural food source like the Chinese oak silk moth pupae. The molecular mechanisms underlying the nutritive impact of the artificial diet on A. chinensis health are unclear. So we utilized transcriptome information to better understand the impact of the artificial diet on A. chinensis at the molecular level.
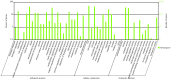
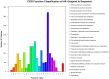
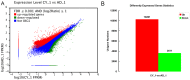
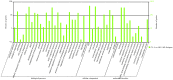
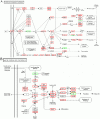
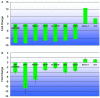
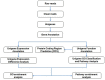
Methodology/principal findings: Illumina HiSeq2000 was used to sequence 4.79 and 4.70 Gb of the transcriptome from pupae-fed and artificial diet-fed A. chinensis libraries, respectively, and a de novo transcriptome assembly was performed (Trinity short read assembler). This resulted in 112,029 and 98,724 contigs, clustered into 54,083 and 54,169 unigenes for pupae-fed and diet-fed A. chinensis, respectively. Unigenes from each sample's assembly underwent sequence splicing and redundancy removal to acquire non-redundant unigenes. We obtained 55,189 unigenes of A. chinensis, including 12,046 distinct clusters and 43,143 distinct singletons. Unigene sequences were aligned by BLASTx to nr, Swiss-Prot, KEGG and COG (E-value <10(-5)), and further aligned by BLASTn to nt (E-value <10(-5)), retrieving proteins of highest sequence similarity with the given unigenes along with their protein functional annotations. Totally, 22,964, 7,898, 18,069, 15,416, 8,066 and 5,341 unigenes were annotated in nr, nt, Swiss-Prot, KEGG, COG and GO, respectively. We compared gene expression variations and found thousands of genes were differentially expressed between pupae-fed and diet-fed A. chinensis.
Conclusions/significance: Our study provides abundant genomic data and offers comprehensive sequence information for studying A. chinensis. Additionally, the physiological roles of the differentially expressed genes enable us to predict effects of some dietary ingredients and subsequently propose formulation improvements to artificial diets.
Conflict of interest statement
Figures

Similar articles
-
Performance and Cost Comparisons for Continuous Rearing of Arma chinensis (Hemiptera: Pentatomidae: Asopinae) on a Zoophytogenous Artificial Diet and a Secondary Prey.J Econ Entomol. 2015 Apr;108(2):454-61. doi: 10.1093/jee/tov024. Epub 2015 Feb 13. J Econ Entomol. 2015. PMID: 26470156
-
Differential Proteomics Analysis Unraveled Mechanisms of Arma chinensis Responding to Improved Artificial Diet.Insects. 2022 Jul 2;13(7):605. doi: 10.3390/insects13070605. Insects. 2022. PMID: 35886781 Free PMC article.
-
De novo assembly and characterization of the global transcriptome for Rhyacionia leptotubula using Illumina paired-end sequencing.PLoS One. 2013 Nov 21;8(11):e81096. doi: 10.1371/journal.pone.0081096. eCollection 2013. PLoS One. 2013. PMID: 24278383 Free PMC article.
-
Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis.PLoS One. 2014 Jan 17;9(1):e86436. doi: 10.1371/journal.pone.0086436. eCollection 2014. PLoS One. 2014. PMID: 24466095 Free PMC article.
-
Population Genomics Provides Insights Into Genomic Features of Inbreeding Depression in Arma Chinensis.Evol Appl. 2025 Jun 8;18(6):e70107. doi: 10.1111/eva.70107. eCollection 2025 Jun. Evol Appl. 2025. PMID: 40495933 Free PMC article. Review.
Cited by
-
Genomic insight into diet adaptation in the biological control agent Cryptolaemus montrouzieri.BMC Genomics. 2021 Feb 25;22(1):135. doi: 10.1186/s12864-021-07442-3. BMC Genomics. 2021. PMID: 33632122 Free PMC article.
-
De novo construction of a transcriptome for the stink bug crop pest Chinavia impicticornis during late development.GigaByte. 2020 Dec 31;2020:gigabyte11. doi: 10.46471/gigabyte.11. eCollection 2020. GigaByte. 2020. PMID: 36824599 Free PMC article.
-
Transcriptome sequencing of Coccinella septempunctata adults (Coleoptera: Coccinellidae) feeding on artificial diet and Aphis craccivora.PLoS One. 2020 Aug 17;15(8):e0236249. doi: 10.1371/journal.pone.0236249. eCollection 2020. PLoS One. 2020. PMID: 32804964 Free PMC article.
-
The transcriptome analysis of Protaetia brevitarsis Lewis larvae.PLoS One. 2019 Mar 21;14(3):e0214001. doi: 10.1371/journal.pone.0214001. eCollection 2019. PLoS One. 2019. PMID: 30897120 Free PMC article.
-
Changes in life history parameters and transcriptome profile of Serangium japonicum associated with feeding on natural prey (Bemisia tabaci) and alternate host (Corcyra cephalonica eggs).BMC Genomics. 2023 Mar 14;24(1):112. doi: 10.1186/s12864-023-09182-y. BMC Genomics. 2023. PMID: 36918764 Free PMC article.
References
-
- Chai XM, He ZH, Jiang P, Wu ZD, Pan CR, et al. (2000) Studies on natural enemies of Dendrolimus punctatus in Zhejiang Province. Journal of Zhejiang Forestry Science and Technology 20: 1–56.
-
- Chen J, Zhang JP, Zhang JH, Tian YH, Xu ZC, et al. (2007) Study on functional response of Arma chinensis to the adults of Monolepta hieroglyphica. . Natural Enemies of Insects 29: 149–154.
-
- Gao CQ, Wang ZM, Yu EY (1993) Studies on artificial rearing of Arma chinensis Fallou. Journal of Jilin Forestry Science and Technology 2: 16–18.
-
- Gao Z (2010) Studies on Biological Characteristic and Control Condition of Arma chinensis Fallou. Harbin: Heilongjiang University Press. p. 11.
-
- Liang ZP, Zhang XX, Song AD, Peng HY (2006) Biology of Clostera anachoreta and its control methods. Chinese Bulletin of Entomology 43: 147–152.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

