Expression of regulatory platelet microRNAs in patients with sickle cell disease
- PMID: 23593351
- PMCID: PMC3625199
- DOI: 10.1371/journal.pone.0060932
Expression of regulatory platelet microRNAs in patients with sickle cell disease
Abstract
Background: Increased platelet activation in sickle cell disease (SCD) contributes to a state of hypercoagulability and confers a risk of thromboembolic complications. The role for post-transcriptional regulation of the platelet transcriptome by microRNAs (miRNAs) in SCD has not been previously explored. This is the first study to determine whether platelets from SCD exhibit an altered miRNA expression profile.
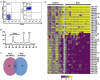
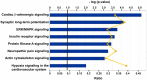
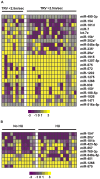
Methods and findings: We analyzed the expression of miRNAs isolated from platelets from a primary cohort (SCD = 19, controls = 10) and a validation cohort (SCD = 7, controls = 7) by hybridizing to the Agilent miRNA microarrays. A dramatic difference in miRNA expression profiles between patients and controls was noted in both cohorts separately. A total of 40 differentially expressed platelet miRNAs were identified as common in both cohorts (p-value 0.05, fold change>2) with 24 miRNAs downregulated. Interestingly, 14 of the 24 downregulated miRNAs were members of three families - miR-329, miR-376 and miR-154 - which localized to the epigenetically regulated, maternally imprinted chromosome 14q32 region. We validated the downregulated miRNAs, miR-376a and miR-409-3p, and an upregulated miR-1225-3p using qRT-PCR. Over-expression of the miR-1225-3p in the Meg01 cells was followed by mRNA expression profiling to identify mRNA targets. This resulted in significant transcriptional repression of 1605 transcripts. A combinatorial approach using Meg01 mRNA expression profiles following miR-1225-3p overexpression, a computational prediction analysis of miRNA target sequences and a previously published set of differentially expressed platelet transcripts from SCD patients, identified three novel platelet mRNA targets: PBXIP1, PLAGL2 and PHF20L1.
Conclusions: We have identified significant differences in functionally active platelet miRNAs in patients with SCD as compared to controls. These data provide an important inventory of differentially expressed miRNAs in SCD patients and an experimental framework for future studies of miRNAs as regulators of biological pathways in platelets.
Conflict of interest statement
Figures


References
-
- Blann AD, Marwah S, Serjeant G, Bareford D, Wright J (2003) Platelet activation and endothelial cell dysfunction in sickle cell disease is unrelated to reduced antioxidant capacity. Blood Coagul Fibrinolysis 14: 255–259. - PubMed
-
- Wun T, Paglieroni T, Rangaswami A, Franklin PH, Welborn J, et al. (1998) Platelet activation in patients with sickle cell disease. Br J Haematol 100: 741–749. - PubMed
-
- Tomer A, Harker LA, Kasey S, Eckman JR (2001) Thrombogenesis in sickle cell disease. J Lab Clin Med 137: 398–407. - PubMed
-
- Mohan JS, Lip GY, Bareford D, Blann AD (2006) Platelet P-selectin and platelet mass, volume and component in sickle cell disease: relationship to genotype. Thromb Res 117: 623–629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

