Critical role of the fusion protein cytoplasmic tail sequence in parainfluenza virus assembly
- PMID: 23593451
- PMCID: PMC3625212
- DOI: 10.1371/journal.pone.0061281
Critical role of the fusion protein cytoplasmic tail sequence in parainfluenza virus assembly
Abstract
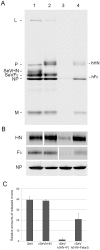
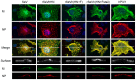
Interactions between viral glycoproteins, matrix protein and nucleocapsid sustain assembly of parainfluenza viruses at the plasma membrane. Although the protein interactions required for virion formation are considered to be highly specific, virions lacking envelope glycoprotein(s) can be produced, thus the molecular interactions driving viral assembly and production are still unclear. Sendai virus (SeV) and human parainfluenza virus type 1 (hPIV1) are highly similar in structure, however, the cytoplasmic tail sequences of the envelope glycoproteins (HN and F) are relatively less conserved. To unveil the specific role of the envelope glycoproteins in viral assembly, we created chimeric SeVs whose HN (rSeVhHN) or HN and F (rSeVh(HN+F)) were replaced with those of hPIV1. rSeVhHN grew as efficiently as wt SeV or hPIV1, suggesting that the sequence difference in HN does not have a significant impact on SeV replication and virion production. In sharp contrast, the growth of rSeVh(HN+F) was significantly impaired compared to rSeVhHN. rSeVh(HN+Fstail) which expresses a chimeric hPIV1 F with the SeV cytoplasmic tail sequence grew similar to wt SeV or rSeVhHN. Further analysis indicated that the F cytoplasmic tail plays a critical role in cell surface expression/accumulation of HN and F, as well as NP and M association at the plasma membrane. Trafficking of nucelocapsids in infected cells was not significantly affected by the origin of F, suggesting that F cytoplasmic tail is not involved in intracellular movement. These results demonstrate the role of the F cytoplasmic tail in accumulation of structural components at the plasma membrane assembly sites.
Conflict of interest statement
Figures

Similar articles
-
Mutation of the TYTLE motif in the cytoplasmic tail of the sendai virus fusion protein deeply affects viral assembly and particle production.PLoS One. 2013 Dec 10;8(12):e78074. doi: 10.1371/journal.pone.0078074. eCollection 2013. PLoS One. 2013. PMID: 24339863 Free PMC article.
-
Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5.J Virol. 2002 Sep;76(18):9284-97. doi: 10.1128/jvi.76.18.9284-9297.2002. J Virol. 2002. PMID: 12186912 Free PMC article.
-
A I131V Substitution in the Fusion Glycoprotein of Human Parainfluenza Virus Type 1 Enhances Syncytium Formation and Virus Growth.Biol Pharm Bull. 2019;42(5):827-832. doi: 10.1248/bpb.b18-00714. Biol Pharm Bull. 2019. PMID: 31061326
-
Paramyxovirus membrane fusion: lessons from the F and HN atomic structures.Virology. 2006 Jan 5;344(1):30-7. doi: 10.1016/j.virol.2005.09.007. Virology. 2006. PMID: 16364733 Free PMC article. Review.
-
The cytoplasmic tail of retroviral envelope glycoproteins.Prog Mol Biol Transl Sci. 2015;129:253-84. doi: 10.1016/bs.pmbts.2014.10.009. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595807 Free PMC article. Review.
Cited by
-
Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins.PLoS Pathog. 2015 Mar 17;11(3):e1004739. doi: 10.1371/journal.ppat.1004739. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25782006 Free PMC article.
-
Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production.Viruses. 2014 Aug 7;6(8):3019-54. doi: 10.3390/v6083019. Viruses. 2014. PMID: 25105277 Free PMC article. Review.
-
Cholesterol reducing agents inhibit assembly of type I parainfluenza viruses.Virology. 2017 Jan 15;501:127-135. doi: 10.1016/j.virol.2016.11.011. Epub 2016 Dec 2. Virology. 2017. PMID: 27915128 Free PMC article.
-
Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody.Front Microbiol. 2014 May 13;5:208. doi: 10.3389/fmicb.2014.00208. eCollection 2014. Front Microbiol. 2014. PMID: 24860558 Free PMC article.
-
Interaction of Human Parainfluenza Virus Type 3 Nucleoprotein with Matrix Protein Mediates Internal Viral Protein Assembly.J Virol. 2015 Dec 9;90(5):2306-15. doi: 10.1128/JVI.02324-15. J Virol. 2015. PMID: 26656716 Free PMC article.
References
-
- Takimoto T, Portner A (2004) Molecular mechanism of paramyxovirus budding. Virus Res 106: 133–145. - PubMed
-
- Lamb RA, Parks GD (2007) Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edition ed. Philadelphia, PA: Lippincott Williams & Wilkins. pp. 1449–1496.
-
- Ali A, Nayak DP (2000) Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276: 289–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

