DNA-intercalators causing rapid re-expression of methylated and silenced genes in cancer cells
- PMID: 23593653
- PMCID: PMC3712575
- DOI: 10.18632/oncotarget.863
DNA-intercalators causing rapid re-expression of methylated and silenced genes in cancer cells
Abstract
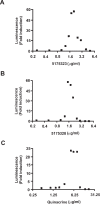
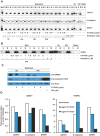
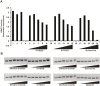
Epigenetic inactivation of tumor-suppressor and other regulatory genes plays a critical role in carcinogenesis. Transcriptional silencing is often maintained by DNA methyl transferase (DNMT)-mediated hypermethylation of CpG islands in promoter DNA. Nucleoside analogs including azacytidine and decitabine have been used to inhibit DNMT and re-activate genes, and are clinically used. Their shortcomings include a short half-life and a slow onset of action due to required nucleotide incorporation during DNA replication, which may limit clinical utility. It might be useful to begin to identify lead compounds having novel properties, specifically distinct and fast-acting gene desilencing. We previously identified chemicals augmenting gene expression in multiple reporter systems. We now report that a subset of these compounds that includes quinacrine re-expresses epigenetically silenced genes implicated in carcinogenesis. p16, TFPI2, the cadherins E-cadherin and CDH13, and the secreted frizzle-related proteins (SFRPs) SFRP1 and SFRP5 were desilenced in cancer cell lines. These lead compounds were fast-acting: re-expression occurred by 12-24 hours. Reactivation of silenced genes was accompanied by depletion of DNMT1 at the promoters of activated genes and demethylation of DNA. A model compound, 5175328, induced changes more rapidly than decitabine. These gene desilencing agents belonged to a class of acridine compounds, intercalated into DNA, and inhibited DNMT1 activity in vitro. Although to define the mechanism would be outside the scope of this initial report, this class may re-activate silenced genes in part by intercalating into DNA and subsequently inhibiting full DNMT1 activity. Rapid mechanisms for chemical desilencing of methylated genes therefore exist.
Figures


Similar articles
-
Activation of silenced tumor suppressor genes in prostate cancer cells by a novel energy restriction-mimetic agent.Prostate. 2012 Dec 1;72(16):1767-78. doi: 10.1002/pros.22530. Epub 2012 Apr 26. Prostate. 2012. PMID: 22539223 Free PMC article.
-
A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation.Cancer Res. 2009 May 15;69(10):4277-85. doi: 10.1158/0008-5472.CAN-08-3669. Epub 2009 May 5. Cancer Res. 2009. Retraction in: Cancer Res. 2023 Dec 15;83(24):4180. doi: 10.1158/0008-5472.CAN-23-2366. PMID: 19417133 Free PMC article. Retracted.
-
Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells.Hum Pathol. 2006 Mar;37(3):298-311. doi: 10.1016/j.humpath.2005.10.013. Hum Pathol. 2006. PMID: 16613325
-
DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer.Curr Cancer Drug Targets. 2013 May;13(4):379-99. doi: 10.2174/15680096113139990077. Curr Cancer Drug Targets. 2013. PMID: 23517596 Review.
-
Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker.Drug Resist Updat. 2004 Aug-Oct;7(4-5):267-78. doi: 10.1016/j.drup.2004.06.005. Drug Resist Updat. 2004. PMID: 15533764 Review.
Cited by
-
Epigenetic repression of phosphatidylethanolamine N-methyltransferase (PEMT) in BRCA1-mutated breast cancer.Oncotarget. 2014 Mar 15;5(5):1315-25. doi: 10.18632/oncotarget.1800. Oncotarget. 2014. PMID: 24675476 Free PMC article.
-
Dietary Flavones as Dual Inhibitors of DNA Methyltransferases and Histone Methyltransferases.PLoS One. 2016 Sep 22;11(9):e0162956. doi: 10.1371/journal.pone.0162956. eCollection 2016. PLoS One. 2016. PMID: 27658199 Free PMC article.
-
Concerted changes in transcriptional regulation of genes involved in DNA methylation, demethylation, and folate-mediated one-carbon metabolism pathways in the NCI-60 cancer cell line panel in response to cancer drug treatment.Clin Epigenetics. 2016 Jun 24;8:73. doi: 10.1186/s13148-016-0240-3. eCollection 2016. Clin Epigenetics. 2016. PMID: 27347216 Free PMC article.
-
DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge.Biomolecules. 2017 Jan 5;7(1):3. doi: 10.3390/biom7010003. Biomolecules. 2017. PMID: 28067760 Free PMC article. Review.
-
Inhibitors of DNA Methylation.Adv Exp Med Biol. 2022;1389:471-513. doi: 10.1007/978-3-031-11454-0_17. Adv Exp Med Biol. 2022. PMID: 36350520 Review.
References
-
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. - PubMed
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
-
- Cohen DE, Melton D. Turning straw into gold: directing cell fate for regenerative medicine. Nat Rev Genet. 12(4):243–252. - PubMed
-
- Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

