Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance
- PMID: 23594083
- PMCID: PMC3884027
- DOI: 10.1021/cb400100g
Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance
Abstract
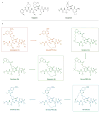
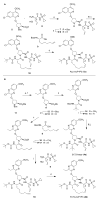
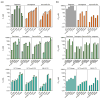
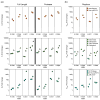
The hepatitis C virus (HCV) infects an estimated 150 million people worldwide and is the major cause of viral hepatitis, cirrhosis, and liver cancer. The available antiviral therapies, which include PEGylated interferon, ribavirin, and one of the HCV NS3/4A protease inhibitors telaprevir or boceprevir, are ineffective for some patients and cause severe side effects. More potent NS3/4A protease inhibitors are in clinical development, but the long-term effectiveness of these drugs is challenged by the development of drug resistance. Here, we investigated the role of macrocycles in the susceptibility of NS3/4A protease inhibitors to drug resistance in asunaprevir, danoprevir, vaniprevir, and MK-5172, with similar core structures but varied P2 moieties and macrocyclizations. Linear and macrocyclic analogues of these drugs were designed, synthesized, and tested against wild-type and drug-resistant variants R155K, V36M/R155K, A156T, and D168A in enzymatic and antiviral assays. Macrocyclic inhibitors were generally more potent, but the location of the macrocycle was critical for retaining activity against drug-resistant variants: the P1-P3 macrocyclic inhibitors were less susceptible to drug resistance than the linear and P2-P4 macrocyclic analogues. In addition, the heterocyclic moiety at P2 largely determined the inhibitor resistance profile, susceptibility to drug resistance, and the extent of modulation by the helicase domain. Our findings suggest that to design robust inhibitors that retain potency to drug-resistant NS3/4A protease variants, inhibitors should combine P1-P3 macrocycles with flexible P2 moieties that optimally contact with the invariable catalytic triad of this enzyme.
Figures

Similar articles
-
The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors.PLoS Pathog. 2012;8(7):e1002832. doi: 10.1371/journal.ppat.1002832. Epub 2012 Jul 26. PLoS Pathog. 2012. PMID: 22910833 Free PMC article.
-
Computational study on the drug resistance mechanism against HCV NS3/4A protease inhibitors vaniprevir and MK-5172 by the combination use of molecular dynamics simulation, residue interaction network, and substrate envelope analysis.J Chem Inf Model. 2014 Feb 24;54(2):621-33. doi: 10.1021/ci400060j. Epub 2013 Jun 28. J Chem Inf Model. 2014. PMID: 23745769
-
Molecular docking investigation of the binding interactions of macrocyclic inhibitors with HCV NS3 protease and its mutants (R155K, D168A and A156V).Protein J. 2014 Feb;33(1):32-47. doi: 10.1007/s10930-013-9538-6. Protein J. 2014. PMID: 24374429
-
Macrocyclic Hepatitis C Virus NS3/4A Protease Inhibitors: An Overview of Medicinal Chemistry.Curr Med Chem. 2016;23(29):3404-3447. doi: 10.2174/0929867323666160510122525. Curr Med Chem. 2016. PMID: 27160539 Review.
-
Advances in the development of macrocyclic inhibitors of hepatitis C virus NS3-4A protease.Curr Top Med Chem. 2010;10(14):1403-22. doi: 10.2174/156802610792232051. Curr Top Med Chem. 2010. PMID: 20536420 Review.
Cited by
-
Viral proteases: Structure, mechanism and inhibition.Enzymes. 2021;50:301-333. doi: 10.1016/bs.enz.2021.09.004. Epub 2021 Nov 17. Enzymes. 2021. PMID: 34861941 Free PMC article.
-
Elucidating the Substrate Envelope of Enterovirus 68-3C Protease: Structural Basis of Specificity and Potential Resistance.Viruses. 2024 Sep 5;16(9):1419. doi: 10.3390/v16091419. Viruses. 2024. PMID: 39339895 Free PMC article.
-
Exploring Macrocyclic Chemical Space: Strategies and Technologies for Drug Discovery.Pharmaceuticals (Basel). 2025 Apr 24;18(5):617. doi: 10.3390/ph18050617. Pharmaceuticals (Basel). 2025. PMID: 40430438 Free PMC article. Review.
-
An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease.J Biomol Struct Dyn. 2021 Aug;39(12):4304-4315. doi: 10.1080/07391102.2020.1776158. Epub 2020 Jun 16. J Biomol Struct Dyn. 2021. PMID: 32544024 Free PMC article.
-
Improving Viral Protease Inhibitors to Counter Drug Resistance.Trends Microbiol. 2016 Jul;24(7):547-557. doi: 10.1016/j.tim.2016.03.010. Epub 2016 Apr 15. Trends Microbiol. 2016. PMID: 27090931 Free PMC article. Review.
References
-
- World Health Organization (WHO) [Accessed February 2013];Hepatitis C, Fact Sheet No 164. 2012 Jul; http://www.who.int/mediacentre/factsheets/fs164/en/index.html.
-
- McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, Alam J, Muir AJ. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. - PubMed
-
- Hézode C, Forestier N, Dusheiko G, Ferenci P, Pol S, Goeser T, Bronowicki JP, Bourlière M, Gharakhanian S, Bengtsson L, McNair L, George S, Kieffer T, Kwong A, Kauffman RS, Alam J, Pawlotsky JM, Zeuzem S. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360:1839–1850. - PubMed
-
- Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N, Rossaro L, Anderson FH, Jacobson IM, Rubin R, Koury K, Pedicone LD, Brass CA, Chaudhri E, Albrecht JK. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376:705–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

