A new simple method for introducing an unmarked mutation into a large gene of non-competent Gram-negative bacteria by FLP/FRT recombination
- PMID: 23594401
- PMCID: PMC3654948
- DOI: 10.1186/1471-2180-13-86
A new simple method for introducing an unmarked mutation into a large gene of non-competent Gram-negative bacteria by FLP/FRT recombination
Abstract
Background: For the disruption of a target gene in molecular microbiology, unmarked mutagenesis is preferable to marked mutagenesis because the former method raises no concern about the polar effect and leaves no selection marker. In contrast to naturally competent bacteria, there is no useful method for introducing an unmarked mutation into a large gene of non-competent bacteria. Nevertheless, large genes encoding huge proteins exist in diverse bacteria and are interesting and important for physiology and potential applications. Here we present a new method for introducing an unmarked mutation into such large genes of non-competent Gram-negative bacteria.
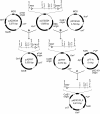
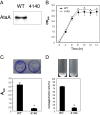
Results: Two gene replacement plasmids, pJQFRT and pKFRT/FLP, were constructed to apply the FLP/FRT recombination system to introduce an unmarked mutation into a large gene of non-competent Gram-negative bacteria. In our methodology, pJQFRT and pKFRT/FLP are integrated into the upstream and the downstream regions of a target gene, respectively, through homologous recombination. The resultant mutant has antibiotic resistance markers, the sacB counter-selection marker, flp recombinase under the control of the tetR regulator, and identical FRT sites sandwiching the target gene and the markers on its chromosome. By inducing the expression of flp recombinase, the target gene is completely deleted together with the other genes derived from the integrated plasmids, resulting in the generation of an unmarked mutation. By this method, we constructed an unmarked mutant of ataA, which encodes the huge trimeric autotransporter adhesin (3,630 aa), in a non-competent Gram-negative bacterium, Acinetobacter sp. Tol 5. The unmarked ataA mutant showed the same growth rate as wild type Tol 5, but lost the adhesive properties of Tol 5, similar to the transposon-inserted mutant of ataA that we generated previously.
Conclusions: The feasibility of our methodology was evidenced by the construction of an unmarked ataA mutant in the Tol 5 strain. Since FLP/FRT recombination can excise a long region of DNA exceeding 100 kb, our method has the potential to selectively disrupt much larger genes or longer regions of gene clusters than ataA. Our methodology allows the straightforward and efficient introduction of an unmarked mutation into a large gene or gene cluster of non-enterobacterial Gram-negative bacteria.
Figures



References
-
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. doi: 10.1038/nbt1183-784. - DOI
-
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

