Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B
- PMID: 23594992
- PMCID: PMC3737343
- DOI: 10.4161/rna.24282
Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B
Abstract
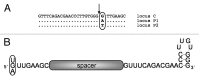
To fend off foreign genetic elements, prokaryotes have developed several defense systems. The most recently discovered defense system, CRISPR/Cas, is sequence-specific, adaptive and heritable. The two central components of this system are the Cas proteins and the CRISPR RNA. The latter consists of repeat sequences that are interspersed with spacer sequences. The CRISPR locus is transcribed into a precursor RNA that is subsequently processed into short crRNAs. CRISPR/Cas systems have been identified in bacteria and archaea, and data show that many variations of this system exist. We analyzed the requirements for a successful defense reaction in the halophilic archaeon Haloferax volcanii. Haloferax encodes a CRISPR/Cas system of the I-B subtype, about which very little is known. Analysis of the mature crRNAs revealed that they contain a spacer as their central element, which is preceded by an eight-nucleotide-long 5' handle that originates from the upstream repeat. The repeat sequences have the potential to fold into a minimal stem loop. Sequencing of the crRNA population indicated that not all of the spacers that are encoded by the three CRISPR loci are present in the same abundance. By challenging Haloferax with an invader plasmid, we demonstrated that the interaction of the crRNA with the invader DNA requires a 10-nucleotide-long seed sequence. In addition, we found that not all of the crRNAs from the three CRISPR loci are effective at triggering the degradation of invader plasmids. The interference does not seem to be influenced by the copy number of the invader plasmid.
Keywords: CRISPR/Cas; Haloferax volcanii; PAM; archaea; crRNA; seed sequence.
Figures


Similar articles
-
An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein.J Biol Chem. 2015 Feb 13;290(7):4192-201. doi: 10.1074/jbc.M114.617506. Epub 2014 Dec 15. J Biol Chem. 2015. PMID: 25512373 Free PMC article.
-
A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii.J Biol Chem. 2014 Mar 7;289(10):7164-7177. doi: 10.1074/jbc.M113.508184. Epub 2014 Jan 23. J Biol Chem. 2014. PMID: 24459147 Free PMC article.
-
Requirements for a successful defence reaction by the CRISPR-Cas subtype I-B system.Biochem Soc Trans. 2013 Dec;41(6):1444-8. doi: 10.1042/BST20130098. Biochem Soc Trans. 2013. PMID: 24256235 Review.
-
Gene Repression in Haloarchaea Using the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas I-B System.J Biol Chem. 2016 Jul 15;291(29):15226-42. doi: 10.1074/jbc.M116.724062. Epub 2016 May 16. J Biol Chem. 2016. PMID: 27226589 Free PMC article.
-
The Adaptive Immune System of Haloferax volcanii.Life (Basel). 2015 Feb 16;5(1):521-37. doi: 10.3390/life5010521. Life (Basel). 2015. PMID: 25692903 Free PMC article. Review.
Cited by
-
Genomic insights into phage-host interaction in the deep-sea chemolithoautotrophic Campylobacterota, Nitratiruptor.ISME Commun. 2022 Nov 1;2(1):108. doi: 10.1038/s43705-022-00194-5. ISME Commun. 2022. PMID: 37938718 Free PMC article.
-
DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus.Nucleic Acids Res. 2015 Dec 2;43(21):10353-63. doi: 10.1093/nar/gkv1140. Epub 2015 Oct 30. Nucleic Acids Res. 2015. PMID: 26519471 Free PMC article.
-
Special focus CRISPR-Cas.RNA Biol. 2013 May;10(5):655-8. doi: 10.4161/rna.24687. RNA Biol. 2013. PMID: 23872677 Free PMC article. No abstract available.
-
Developing a genetic manipulation system for the Antarctic archaeon, Halorubrum lacusprofundi: investigating acetamidase gene function.Sci Rep. 2016 Oct 6;6:34639. doi: 10.1038/srep34639. Sci Rep. 2016. PMID: 27708407 Free PMC article.
-
Repeat modularity as a beneficial property of multiple CRISPR-Cas systems.RNA Biol. 2019 Apr;16(4):585-587. doi: 10.1080/15476286.2018.1474073. Epub 2018 Aug 10. RNA Biol. 2019. PMID: 29923454 Free PMC article.
References
-
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Review: Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;2011:7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
