Identification of a genomic region containing a novel promoter resistant to glucose repression and over-expression of β-glucosidase gene in Hypocrea orientalis EU7-22
- PMID: 23594998
- PMCID: PMC3645756
- DOI: 10.3390/ijms14048479
Identification of a genomic region containing a novel promoter resistant to glucose repression and over-expression of β-glucosidase gene in Hypocrea orientalis EU7-22
Abstract
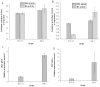
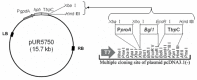
A high concentration of glucose in the medium could greatly inhibit the expression of cellulase in filamentous fungi. The aspartic protease from fungus Hypocrea orientalis EU7-22 could efficiently express under both induction condition and glucose repression condition. Based on the sequence of structure gene of aspartic protease, the upstream sequence harboring the putative promoter proA for driving the expression of aspartic protease was obtained by genome walking. The upstream sequence contained the typical promoter motifs "TATA" and "CAAT". The β-glucosidase gene (Bgl1) from H. orientalis was cloned and recombined with promoter proA and terminator trpC. The expression cassette was ligated to the binary vector to form pUR5750-Bgl1, and then transferred into the host strain EU7-22 via Agrobacterium tumefaciens mediated transformation (ATMT), using hygromycin B resistance gene as the screening marker. Four transformants Bgl-1, Bgl-2, Bgl-3 and Bgl-4 were screened. Compared with the host strain EU7-22, the enzyme activities of filter paper (FPA) and β-glucosidase (BG) of transformant Bgl-2 increased by 10.6% and 19.1% under induction condition, respectively. The FPA and BG activities were enhanced by 22.2% and 700% under 2% glucose repression condition, respectively, compared with the host strain. The results showed that the putative promoter proA has successfully driven the over-expression of Bgl1 gene in H. orientalis under glucose repression condition.
Figures
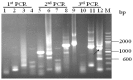

 ”) involved in carbon, nitrogen, and pH regulations, respectively. Arrow underline the sequences indicated the reverse primer SP3. M = A or C; S = G or C; Y = C or T; R = A or G; H = A, C or T.
”) involved in carbon, nitrogen, and pH regulations, respectively. Arrow underline the sequences indicated the reverse primer SP3. M = A or C; S = G or C; Y = C or T; R = A or G; H = A, C or T.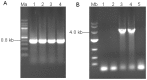

Similar articles
-
Inactivation kinetics and conformation change of Hypocrea orientalis β-glucosidase with guanidine hydrochloride.J Biosci Bioeng. 2017 Aug;124(2):143-149. doi: 10.1016/j.jbiosc.2017.03.006. Epub 2017 Apr 11. J Biosci Bioeng. 2017. PMID: 28410898
-
The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellulose-inducible beta-glucosidase involved in cellulase induction by sophorose.Mol Microbiol. 1995 May;16(4):687-97. doi: 10.1111/j.1365-2958.1995.tb02430.x. Mol Microbiol. 1995. PMID: 7476163
-
Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina.Eukaryot Cell. 2006 Mar;5(3):447-56. doi: 10.1128/EC.5.3.447-456.2006. Eukaryot Cell. 2006. PMID: 16524900 Free PMC article.
-
[Mechanisms and regulation of enzymatic hydrolysis of cellulose in filamentous fungi: classical cases and new models].Rev Iberoam Micol. 2015 Jan-Mar;32(1):1-12. doi: 10.1016/j.riam.2013.10.009. Epub 2014 Mar 7. Rev Iberoam Micol. 2015. PMID: 24607657 Review. Spanish.
-
[Transcriptional regulation of cellulases and hemicellulases gene in Hypocrea jecorina--a review].Wei Sheng Wu Xue Bao. 2010 Nov;50(11):1431-7. Wei Sheng Wu Xue Bao. 2010. PMID: 21268886 Review. Chinese.
Cited by
-
Improvement in xylooligosaccharides production by knockout of the β-xyl1 gene in Trichoderma orientalis EU7-22.3 Biotech. 2018 Jan;8(1):26. doi: 10.1007/s13205-017-1041-x. Epub 2017 Dec 20. 3 Biotech. 2018. PMID: 29279819 Free PMC article.
-
Enhancing Cellulase and Hemicellulase Production in Trichoderma orientalis EU7-22 via Knockout of the creA.Mol Biotechnol. 2018 Jan;60(1):55-61. doi: 10.1007/s12033-017-0046-3. Mol Biotechnol. 2018. PMID: 29214500
References
-
- Kamm B., Kamm M. Biorefineries–Multi product processes. Adv. Biochem. Eng. Biotechnol. 2007;105:175–204. - PubMed
-
- Kumar R., Singh S., Singh O.V. Bioconversion of lignocellulosic biomass: Biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 2008;35:377–391. - PubMed
-
- Kovács K., Szakacs G., Zacchi G. Comparative enzymatic hydrolysis of pretreated spruce by supernatants, whole fermentation broths and washed mycelia of Trichoderma reesei and Trichoderma atroviride. Bioresour. Technol. 2009;100:1350–1357. - PubMed
-
- Sipos B., Benko Z., Dienes D., Réczey K., Viikari L., Siika-aho M. Characterisation of specific activities and hydrolytic properties of cell-wall degrading enzymes produced by Trichoderma reesei Rut C30 on different carbon sources. Appl. Biochem. Biotechnol. 2010;161:347–364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

