Long non-coding RNAs as targets for cytosine methylation
- PMID: 23595112
- PMCID: PMC4111728
- DOI: 10.4161/rna.24454
Long non-coding RNAs as targets for cytosine methylation
Abstract
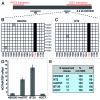
Post-synthetic modifications of nucleic acids have long been known to affect their functional and structural properties. For instance, numerous different chemical modifications modulate the structural organization, stability or translation efficiency of tRNAs and rRNAs. In contrast, little is known about modifications of poly(A)RNAs. Here, we demonstrate for the first time that the two well-studied regulatory long non-coding RNAs HOTAIR and XIST are targets of site-specific cytosine methylation. In both XIST and HOTAIR, we found methylated cytosines located within or near functionally important regions that are known to mediate interaction with chromatin-associated protein complexes. We show that cytosine methylation in the XIST A structure strongly affects binding to the chromatin-modifying complex PRC2 in vitro. These results suggest that cytosine methylation may serve as a general strategy to regulate the function of long non-coding RNAs.
Keywords: 5-methylcytosine; HOTAIR RNA; RNA methylation; XIST RNA; chromatin; long non-coding RNA; polycomb repressive complex 2.
Figures


References
-
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–32. - PubMed
-
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas: Landes Bioscience, 2009:1-18.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
