Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types
- PMID: 23595227
- PMCID: PMC3698507
- DOI: 10.1101/gr.144840.112
Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types
Abstract
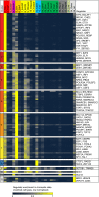
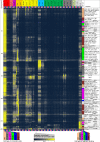
The regions bound by sequence-specific transcription factors can be highly variable across different cell types despite the static nature of the underlying genome sequence. This has been partly attributed to changes in chromatin accessibility, but a systematic picture has been hindered by the lack of large-scale data sets. Here, we use 456 binding experiments for 119 regulators and 84 chromatin maps generated by the ENCODE in six human cell types, and relate those to a global map of regulatory motif instances for these factors. We find specific and robust chromatin state preferences for each regulator beyond the previously reported open-chromatin association, suggesting a much richer chromatin landscape beyond simple accessibility. The preferentially bound chromatin states of regulators were enriched for sequence motifs of regulators relative to all states, suggesting that these preferences are at least partly encoded by the genomic sequence. Relative to all regions bound by a regulator, however, regulatory motifs were surprisingly depleted in the regulator's preferentially bound states, suggesting additional non-sequence-specific binding beyond the level predicted by the regulatory motifs. Such permissive binding was largely restricted to open-chromatin regions showing histone modification marks characteristic of active enhancer and promoter regions, whereas open-chromatin regions lacking such marks did not show permissive binding. Lastly, the vast majority of cobinding of regulator pairs is predicted by the chromatin state preferences of individual regulators. Overall, our results suggest a joint role of sequence motifs and specific chromatin states beyond mere accessibility in mediating regulator binding dynamics across different cell types.
Figures



References
-
- Applegate DL, Bixby RE, Chvatal V, Cook WJ 2006. The traveling salesman problem: A computational study. Princeton University Press, Princeton, NJ
-
- Bar-Joseph Z, Gifford DK, Jaakkola TS 2001. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics 17: S22. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Biedl T, Brejová B, Demaine ED, Hamel AM, Vinar T 2001. Optimal arrangement of leaves in the tree representing hierarchical clustering of gene expression data. Tech. Rep. 2001, Department of Computer Science, University of Waterloo, Ontario, Canada, p. 14
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
