A self-updating road map of The Cancer Genome Atlas
- PMID: 23595662
- PMCID: PMC3654710
- DOI: 10.1093/bioinformatics/btt141
A self-updating road map of The Cancer Genome Atlas
Abstract
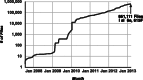
Motivation: Since 2011, The Cancer Genome Atlas' (TCGA) files have been accessible through HTTP from a public site, creating entirely new possibilities for cancer informatics by enhancing data discovery and retrieval. Significantly, these enhancements enable the reporting of analysis results that can be fully traced to and reproduced using their source data. However, to realize this possibility, a continually updated road map of files in the TCGA is required. Creation of such a road map represents a significant data modeling challenge, due to the size and fluidity of this resource: each of the 33 cancer types is instantiated in only partially overlapping sets of analytical platforms, while the number of data files available doubles approximately every 7 months.
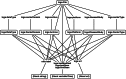
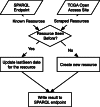
Results: We developed an engine to index and annotate the TCGA files, relying exclusively on third-generation web technologies (Web 3.0). Specifically, this engine uses JavaScript in conjunction with the World Wide Web Consortium's (W3C) Resource Description Framework (RDF), and SPARQL, the query language for RDF, to capture metadata of files in the TCGA open-access HTTP directory. The resulting index may be queried using SPARQL, and enables file-level provenance annotations as well as discovery of arbitrary subsets of files, based on their metadata, using web standard languages. In turn, these abilities enhance the reproducibility and distribution of novel results delivered as elements of a web-based computational ecosystem. The development of the TCGA Roadmap engine was found to provide specific clues about how biomedical big data initiatives should be exposed as public resources for exploratory analysis, data mining and reproducible research. These specific design elements align with the concept of knowledge reengineering and represent a sharp departure from top-down approaches in grid initiatives such as CaBIG. They also present a much more interoperable and reproducible alternative to the still pervasive use of data portals.
Availability: A prepared dashboard, including links to source code and a SPARQL endpoint, is available at http://bit.ly/TCGARoadmap. A video tutorial is available at http://bit.ly/TCGARoadmapTutorial.
Contact: robbinsd@uab.edu.
Figures


References
-
- Baggerly KA, Berry DA. Reproducible research. AMSTAT News. 2011:pp. 16–17.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

