Dominant effects of the Huntington's disease HTT CAG repeat length are captured in gene-expression data sets by a continuous analysis mathematical modeling strategy
- PMID: 23595883
- PMCID: PMC3723309
- DOI: 10.1093/hmg/ddt176
Dominant effects of the Huntington's disease HTT CAG repeat length are captured in gene-expression data sets by a continuous analysis mathematical modeling strategy
Abstract
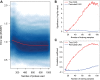
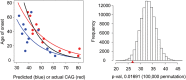
In Huntington's disease (HD), the size of the expanded HTT CAG repeat mutation is the primary driver of the processes that determine age at onset of motor symptoms. However, correlation of cellular biochemical parameters also extends across the normal repeat range, supporting the view that the CAG repeat represents a functional polymorphism with dominant effects determined by the longer allele. A central challenge to defining the functional consequences of this single polymorphism is the difficulty of distinguishing its subtle effects from the multitude of other sources of biological variation. We demonstrate that an analytical approach based upon continuous correlation with CAG size was able to capture the modest (∼21%) contribution of the repeat to the variation in genome-wide gene expression in 107 lymphoblastoid cell lines, with alleles ranging from 15 to 92 CAGs. Furthermore, a mathematical model from an iterative strategy yielded predicted CAG repeat lengths that were significantly positively correlated with true CAG allele size and negatively correlated with age at onset of motor symptoms. Genes negatively correlated with repeat size were also enriched in a set of genes whose expression were CAG-correlated in human HD cerebellum. These findings both reveal the relatively small, but detectable impact of variation in the CAG allele in global data in these peripheral cells and provide a strategy for building multi-dimensional data-driven models of the biological network that drives the HD disease process by continuous analysis across allelic panels of neuronal cells vulnerable to the dominant effects of the HTT CAG repeat.
Figures




References
-
- HDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. - PubMed
-
- Huntington G. On chorea. Med. Surg. Rep. 1872;26:320–321.
-
- Hendricks A.E., Latourelle J.C., Lunetta K.L., Cupples L.A., Wheeler V., MacDonald M.E., Gusella J.F., Myers R.H. Estimating the probability of de novo HD cases from transmissions of expanded penetrant CAG alleles in the Huntington disease gene from male carriers of high normal alleles (27-35 CAG) Am. J. Med. Genet. A. 2009;149A:1375–1381. - PMC - PubMed
-
- Langbehn D.R., Brinkman R.R., Falush D., Paulsen J.S., Hayden M.R. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin. Genet. 2004;65:267–277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

