Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer
- PMID: 23596205
- PMCID: PMC3897241
- DOI: 10.1126/scitranslmed.3006112
Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer
Abstract
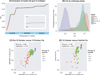
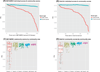
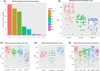
Although molecular prognostics in breast cancer are among the most successful examples of translating genomic analysis to clinical applications, optimal approaches to breast cancer clinical risk prediction remain controversial. The Sage Bionetworks-DREAM Breast Cancer Prognosis Challenge (BCC) is a crowdsourced research study for breast cancer prognostic modeling using genome-scale data. The BCC provided a community of data analysts with a common platform for data access and blinded evaluation of model accuracy in predicting breast cancer survival on the basis of gene expression data, copy number data, and clinical covariates. This approach offered the opportunity to assess whether a crowdsourced community Challenge would generate models of breast cancer prognosis commensurate with or exceeding current best-in-class approaches. The BCC comprised multiple rounds of blinded evaluations on held-out portions of data on 1981 patients, resulting in more than 1400 models submitted as open source code. Participants then retrained their models on the full data set of 1981 samples and submitted up to five models for validation in a newly generated data set of 184 breast cancer patients. Analysis of the BCC results suggests that the best-performing modeling strategy outperformed previously reported methods in blinded evaluations; model performance was consistent across several independent evaluations; and aggregating community-developed models achieved performance on par with the best-performing individual models.
Figures


Comment in
-
Prognostic models: rising to the challenge.Nat Rev Cancer. 2013 Jun;13(6):378. doi: 10.1038/nrc3530. Epub 2013 May 3. Nat Rev Cancer. 2013. PMID: 23640208 No abstract available.
References
-
- Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 2013;132:1133–1145. - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Rijn Mvan de S.S. Jeffrey, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10869–10874. - PMC - PubMed
-
- Vijver van de MJ, He YD, van’t Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

