Observations on the evolution of the melanocortin receptor gene family: distinctive features of the melanocortin-2 receptor
- PMID: 23596380
- PMCID: PMC3622036
- DOI: 10.3389/fnins.2013.00028
Observations on the evolution of the melanocortin receptor gene family: distinctive features of the melanocortin-2 receptor
Abstract
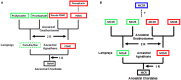
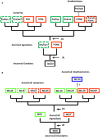
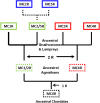
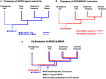
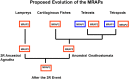
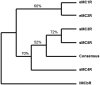
The melanocortin receptors (MCRs) are a gene family in the rhodopsin class of G protein-coupled receptors. Based on the analysis of several metazoan genome databases it appears that the MCRs are only found in chordates. The presence of five genes in the family (i.e., mc1r, mc2r, mc3r, mc4r, mc5r) in representatives of the tetrapods indicates that the gene family is the result of two genome duplication events and one local gene duplication event during the evolution of the chordates. The MCRs are activated by melanocortin ligands (i.e., ACTH, α-MSH, β-MSH, γ-MSH, δ-MSH) which are all derived from the polypeptide hormone/neuropeptide precursor, POMC, and as a result the functional evolution of the MCRs is intimately associated with the co-evolution of POMC endocrine and neuronal circuits. This review will consider the origin of the MCRs, and discuss the evolutionary relationship between MC2R, MC5R, and MC4R. In addition, this review will analyze the functional evolution of the mc2r gene in light of the co-evolution of the MRAP (Melanocortin-2 Receptor Accessory Protein) gene family.
Keywords: ACTH; MC2R; MC5R; MRAP; constructive neutral evolution; evolutionary cell biology; melanocortin receptors; α-MSH.
Figures

Similar articles
-
60 YEARS OF POMC: Melanocortin receptors: evolution of ligand selectivity for melanocortin peptides.J Mol Endocrinol. 2016 May;56(4):T119-33. doi: 10.1530/JME-15-0292. Epub 2016 Jan 20. J Mol Endocrinol. 2016. PMID: 26792827 Review.
-
The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs.Mol Biol Evol. 2004 Mar;21(3):563-79. doi: 10.1093/molbev/msh050. Epub 2003 Dec 23. Mol Biol Evol. 2004. PMID: 14694081
-
Molecular evolution of GPCRs: Melanocortin/melanocortin receptors.J Mol Endocrinol. 2014 Jun;52(3):T29-42. doi: 10.1530/JME-14-0050. J Mol Endocrinol. 2014. PMID: 24868105 Review.
-
Implication of Melanocortin Receptor Genes in the Familial Comorbidity of Type 2 Diabetes and Depression.Int J Mol Sci. 2022 Jul 28;23(15):8350. doi: 10.3390/ijms23158350. Int J Mol Sci. 2022. PMID: 35955479 Free PMC article.
-
Emerging roles of melanocortin receptor accessory proteins (MRAP and MRAP2) in physiology and pathophysiology.Gene. 2020 Oct 5;757:144949. doi: 10.1016/j.gene.2020.144949. Epub 2020 Jul 15. Gene. 2020. PMID: 32679290 Free PMC article. Review.
Cited by
-
Hypothesis and Theory: Revisiting Views on the Co-evolution of the Melanocortin Receptors and the Accessory Proteins, MRAP1 and MRAP2.Front Endocrinol (Lausanne). 2016 Jun 28;7:79. doi: 10.3389/fendo.2016.00079. eCollection 2016. Front Endocrinol (Lausanne). 2016. PMID: 27445982 Free PMC article.
-
Characterization of the chicken melanocortin 5 receptor and its potential role in regulating hepatic glucolipid metabolism.Front Physiol. 2022 Oct 6;13:917712. doi: 10.3389/fphys.2022.917712. eCollection 2022. Front Physiol. 2022. PMID: 36277187 Free PMC article.
-
Functional Characterization of the Internal Symmetry of MRAP2 Antiparallel Homodimer.Front Endocrinol (Lausanne). 2021 Oct 25;12:750797. doi: 10.3389/fendo.2021.750797. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34759891 Free PMC article.
-
Melanocortin receptor accessory proteins in adrenal disease and obesity.Front Neurosci. 2015 Jun 10;9:213. doi: 10.3389/fnins.2015.00213. eCollection 2015. Front Neurosci. 2015. PMID: 26113808 Free PMC article. Review.
-
Melanocortin-5 Receptor: Pharmacology and Its Regulation of Energy Metabolism.Int J Mol Sci. 2022 Aug 5;23(15):8727. doi: 10.3390/ijms23158727. Int J Mol Sci. 2022. PMID: 35955857 Free PMC article. Review.
References
-
- Carroll R. L. (1988). Vertebrate Paleontology and Evolution. New York: Freeman Press
-
- Chung T. T., Webb T. R., Chan L. F., Cooray S. N., Metherell L. A., King P. J., et al. (2008). The majority of ACTH receptor (MC2R) mutations found in familial glucocorticoid deficiency type 1 lead to defective trafficking of the receptor to the cell surface. J. Clin. Endocrinol. Metab. 93, 4948–495410.1210/jc.2008-1744 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

