Hybrid approach for predicting coreceptor used by HIV-1 from its V3 loop amino acid sequence
- PMID: 23596523
- PMCID: PMC3626595
- DOI: 10.1371/journal.pone.0061437
Hybrid approach for predicting coreceptor used by HIV-1 from its V3 loop amino acid sequence
Erratum in
- PLoS One. 2013;8(11). doi:10.1371/annotation/5c57dcdc-e5d9-4999-a7d0-32004427cba5
Abstract
Background: HIV-1 infects the host cell by interacting with the primary receptor CD4 and a coreceptor CCR5 or CXCR4. Maraviroc, a CCR5 antagonist binds to CCR5 receptor. Thus, it is important to identify the coreceptor used by the HIV strains dominating in the patient. In past, a number of experimental assays and in-silico techniques have been developed for predicting the coreceptor tropism. The prediction accuracy of these methods is excellent when predicting CCR5(R5) tropic sequences but is relatively poor for CXCR4(X4) tropic sequences. Therefore, any new method for accurate determination of coreceptor usage would be of paramount importance to the successful management of HIV-infected individuals.
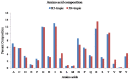
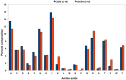
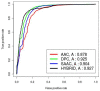
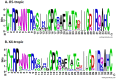
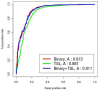
Results: The dataset used in this study comprised 1799 R5-tropic and 598 X4-tropic third variable (V3) sequences of HIV-1. We compared the amino acid composition of both types of V3 sequences and observed that certain types of residues, e.g., Asparagine and Isoleucine, were preferred in R5-tropic sequences whereas residues like Lysine, Arginine, and Tryptophan were preferred in X4-tropic sequences. Initially, Support Vector Machine-based models were developed using amino acid composition, dipeptide composition, and split amino acid composition, which achieved accuracy up to 90%. We used BLAST to discriminate R5- and X4-tropic sequences and correctly predicted 93.16% of R5- and 75.75% of X4-tropic sequences. In order to improve the prediction accuracy, a Hybrid model was developed that achieved 91.66% sensitivity, 81.77% specificity, 89.19% accuracy and 0.72 Matthews Correlation Coefficient. The performance of our models was also evaluated on an independent dataset (256 R5- and 81 X4-tropic sequences) and achieved maximum accuracy of 84.87% with Matthews Correlation Coefficient 0.63.
Conclusion: This study describes a highly efficient method for predicting HIV-1 coreceptor usage from V3 sequences. In order to provide a service to the scientific community, a webserver HIVcoPred was developed (http://www.imtech.res.in/raghava/hivcopred/) for predicting the coreceptor usage.
Conflict of interest statement
Figures

References
-
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, et al. (1996) The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med 2: 1244–1247. - PubMed
-
- Berger EA, Murphy PM, Farber JM (1999) Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 17: 657–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

