Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex
- PMID: 23597049
- PMCID: PMC3640958
- DOI: 10.1186/1744-8069-9-21
Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex
Abstract
Background: Peripheral nerve injury can have long-term consequences including pain-related manifestations, such as hypersensitivity to cutaneous stimuli, as well as affective and cognitive disturbances, suggesting the involvement of supraspinal mechanisms. Changes in brain structure and cortical function associated with many chronic pain conditions have been reported in the prefrontal cortex (PFC). The PFC is implicated in pain-related co-morbidities such as depression, anxiety and impaired emotional decision-making ability. We recently reported that this region is subject to significant epigenetic reprogramming following peripheral nerve injury, and normalization of pain-related structural, functional and epigenetic abnormalities in the PFC are all associated with effective pain reduction. In this study, we used the Spared Nerve Injury (SNI) model of neuropathic pain to test the hypothesis that peripheral nerve injury triggers persistent long-lasting changes in gene expression in the PFC, which alter functional gene networks, thus providing a possible explanation for chronic pain associated behaviors.
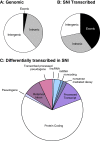
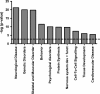
Results: SNI or sham surgery where performed in male CD1 mice at three months of age. Six months after injury, we performed transcriptome-wide sequencing (RNAseq), which revealed 1147 differentially regulated transcripts in the PFC in nerve-injured vs. control mice. Changes in gene expression occurred across a number of functional gene clusters encoding cardinal biological processes as revealed by Ingenuity Pathway Analysis. Significantly altered biological processes included neurological disease, skeletal muscular disorders, behavior, and psychological disorders. Several of the changes detected by RNAseq were validated by RT-QPCR and included transcripts with known roles in chronic pain and/or neuronal plasticity including the NMDA receptor (glutamate receptor, ionotropic, NMDA; grin1), neurite outgrowth (roundabout 3; robo3), gliosis (glial fibrillary acidic protein; gfap), vesicular release (synaptotagmin 2; syt2), and neuronal excitability (voltage-gated sodium channel, type I; scn1a).
Conclusions: This study used an unbiased approach to document long-term alterations in gene expression in the brain following peripheral nerve injury. We propose that these changes are maintained as a memory of an insult that is temporally and spatially distant from the initial injury.
Figures




References
-
- Grace PM, Hurley D, Barratt DT, Tsykin A, Watkins LR, Rolan PE, Hutchinson MR. Harnessing pain heterogeneity and RNA transcriptome to identify blood-based pain biomarkers: a novel correlational study design and bioinformatics approach in a graded chronic constriction injury model. J Neurochem. 2012;122:976–994. doi: 10.1111/j.1471-4159.2012.07833.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

