Gene regulatory networks governing pancreas development
- PMID: 23597482
- PMCID: PMC3645877
- DOI: 10.1016/j.devcel.2013.03.016
Gene regulatory networks governing pancreas development
Abstract
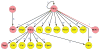
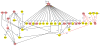
Elucidation of cellular and gene regulatory networks (GRNs) governing organ development will accelerate progress toward tissue replacement. Here, we have compiled reference GRNs underlying pancreas development from data mining that integrates multiple approaches, including mutant analysis, lineage tracing, cell purification, gene expression and enhancer analysis, and biochemical studies of gene regulation. Using established computational tools, we integrated and represented these networks in frameworks that should enhance understanding of the surging output of genomic-scale genetic and epigenetic studies of pancreas development and diseases such as diabetes and pancreatic cancer. We envision similar approaches would be useful for understanding the development of other organs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

