Nitrogenase reduction of carbon-containing compounds
- PMID: 23597875
- PMCID: PMC3714343
- DOI: 10.1016/j.bbabio.2013.04.003
Nitrogenase reduction of carbon-containing compounds
Abstract
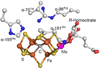
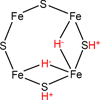
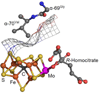
Nitrogenase is an enzyme found in many bacteria and archaea that catalyzes biological dinitrogen fixation, the reduction of N2 to NH3, accounting for the major input of fixed nitrogen into the biogeochemical N cycle. In addition to reducing N2 and protons, nitrogenase can reduce a number of small, non-physiological substrates. Among these alternative substrates are included a wide array of carbon-containing compounds. These compounds have provided unique insights into aspects of the nitrogenase mechanism. Recently, it was shown that carbon monoxide (CO) and carbon dioxide (CO2) can also be reduced by nitrogenase to yield hydrocarbons, opening new insights into the mechanism of small molecule activation and reduction by this complex enzyme as well as providing clues for the design of novel molecular catalysts. This article is part of a Special Issue entitled: Metals in Bioenergetics and Biomimetics Systems.
Keywords: Carbon; Carbon dioxide; Carbon monoxide; Reduction; Substrate.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures


References
-
- Smil V. Enriching the Earth: Fritz Haber, Carl Bosch, and the Transformation of World Food Production. Cambridge, MA: The MIT Press; 2004.
-
- Eady RR. Structure-function relationships of alternative nitrogenases. Chem. Rev. 1996;96:3013–3030. - PubMed
-
- Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem. Rev. 1996;96:2983–3012. - PubMed
-
- Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

