DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development
- PMID: 23598468
- PMCID: PMC3731807
- DOI: 10.1093/carcin/bgt129
DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development
Abstract
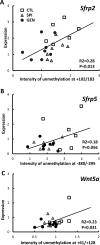
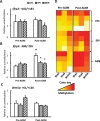
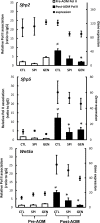
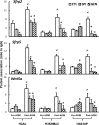
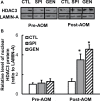
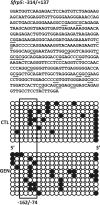
This study aims to elucidate the epigenetic mechanisms by which genistein (GEN) maintains a normal level of WNT genes during colon cancer development. We have reported that soy protein isolate (SPI) and GEN repressed WNT signaling, correlating with the reduction of pre-neoplastic lesions in rat colon. We hypothesized that SPI and GEN induced epigenetic modifications on Sfrp2, Sfrp5 and Wnt5a genes, suppressing their gene expression induced by azoxymethane (AOM), a chemical carcinogen, to the similar level as that of pre-AOM period. We identified that in the post-AOM period, histone H3 acetylation (H3Ac) was downregulated by SPI and GEN at the promoter region of Sfrp2, Sfrp5 and Wnt5a, which paralleled with the reduced binding of RNA polymerase II. Nuclear level of histone deacetylase 3 was enhanced by SPI and GEN. The diets suppressed the trimethylation of histone H3 Lysine 9 (H3K9Me3) and the phosphorylation of histone H3 Serine 10 (H3S10P). Methylation of the specific region of Sfrp2, Sfrp5 and Wnt5a genes was increased by SPI and GEN, which was inversely correlated with the reduction of gene expression. Bisulfite sequencing further confirmed that dietary GEN induced DNA methylation at CpG island of the promoter region of Sfrp5. Importantly, this region includes a fragment that had decreased H3Ac. Here, we present a potential epigenetic mechanism by which dietary GEN controls the responses of WNT genes during carcinogen induction, which involves DNA methylation, histone modifications and their interactions at the regulatory region of gene.
Figures
Similar articles
-
Genistein, a soya isoflavone, prevents azoxymethane-induced up-regulation of WNT/β-catenin signalling and reduces colon pre-neoplasia in rats.Br J Nutr. 2013 Jan 14;109(1):33-42. doi: 10.1017/S0007114512000876. Epub 2012 Apr 3. Br J Nutr. 2013. PMID: 22716201
-
Genistein increases gene expression by demethylation of WNT5a promoter in colon cancer cell line SW1116.Anticancer Res. 2010 Nov;30(11):4537-45. Anticancer Res. 2010. PMID: 21115903
-
Silencing of Wnt5a during colon cancer metastasis involves histone modifications.Epigenetics. 2012 Jun 1;7(6):551-8. doi: 10.4161/epi.20050. Epub 2012 Jun 1. Epigenetics. 2012. PMID: 22522911 Free PMC article.
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
-
Genistein as a regulator of signaling pathways and microRNAs in different types of cancers.Cancer Cell Int. 2021 Jul 21;21(1):388. doi: 10.1186/s12935-021-02091-8. Cancer Cell Int. 2021. PMID: 34289845 Free PMC article. Review.
-
WNT5a in Colorectal Cancer: Research Progress and Challenges.Cancer Manag Res. 2021 Mar 16;13:2483-2498. doi: 10.2147/CMAR.S289819. eCollection 2021. Cancer Manag Res. 2021. PMID: 33758546 Free PMC article. Review.
-
Reduced Representation Bisulfite Sequencing Determination of Distinctive DNA Hypermethylated Genes in the Progression to Colon Cancer in African Americans.Gastroenterol Res Pract. 2016;2016:2102674. doi: 10.1155/2016/2102674. Epub 2016 Sep 1. Gastroenterol Res Pract. 2016. PMID: 27688749 Free PMC article.
-
Effects of Phytoestrogens on the Developing Brain, Gut Microbiota, and Risk for Neurobehavioral Disorders.Front Nutr. 2019 Aug 29;6:142. doi: 10.3389/fnut.2019.00142. eCollection 2019. Front Nutr. 2019. PMID: 31555657 Free PMC article. Review.
-
Epigenetic activities of flavonoids in the prevention and treatment of cancer.Clin Epigenetics. 2015 Jul 10;7(1):64. doi: 10.1186/s13148-015-0095-z. eCollection 2015. Clin Epigenetics. 2015. PMID: 26161152 Free PMC article.
References
-
- Portela A., et al. (2010). Epigenetic modifications and human disease. Nat. Biotechnol., 28, 1057–1068 - PubMed
-
- Esteller M. (2007). Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br. J. Cancer, 96 (suppl.), R26–R30 - PubMed
-
- Kim M.S., et al. (2010). DNA methylation markers in colorectal cancer. Cancer Metastasis Rev., 29, 181–206 - PubMed
-
- Konishi K., et al. (2007). Targeting aberrant chromatin structure in colorectal carcinomas. Cancer J., 13, 49–55 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

