A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts
- PMID: 23599343
- PMCID: PMC3650231
- DOI: 10.1101/gad.213827.113
A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts
Abstract
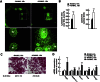
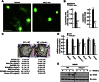
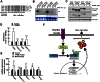
Bone resorption by osteoclasts requires a large number of lysosomes that release proteases in the resorption lacuna. Whether lysosomal biogenesis is a consequence of the action of transcriptional regulators of osteoclast differentiation or is under the control of a different and specific transcriptional pathway remains unknown. We show here, through cell-based assays and cell-specific gene deletion experiments in mice, that the osteoclast differentiation factor RANKL promotes lysosomal biogenesis once osteoclasts are differentiated through the selective activation of TFEB, a member of the MITF/TFE family of transcription factors. This occurs following PKCβ phosphorylation of TFEB on three serine residues located in its last 15 amino acids. This post-translational modification stabilizes and increases the activity of this transcription factor. Supporting these biochemical observations, mice lacking in osteoclasts--either TFEB or PKCβ--show decreased lysosomal gene expression and increased bone mass. Altogether, these results uncover a RANKL-dependent signaling pathway taking place in differentiated osteoclasts and culminating in the activation of TFEB to enhance lysosomal biogenesis-a necessary step for proper bone resorption.
Figures





References
-
- Carralot JP, Kim TK, Lenseigne B, Boese AS, Sommer P, Genovesio A, Brodin P 2009. Automated high-throughput siRNA transfection in raw 264.7 macrophages: A case study for optimization procedure. J Biomol Screen 14: 151–160 - PubMed
-
- Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J 2003. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med 9: 399–406 - PubMed
-
- Chappard D, Palle S, Alexandre C, Vico L, Riffat G 1987. Bone embedding in pure methyl methacrylate at low temperature preserves enzyme activities. Acta Histochem 81: 183–190 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
