Protein kinase C δ increases Kruppel-like factor 4 protein, which drives involucrin gene transcription in differentiating keratinocytes
- PMID: 23599428
- PMCID: PMC3682575
- DOI: 10.1074/jbc.M113.477133
Protein kinase C δ increases Kruppel-like factor 4 protein, which drives involucrin gene transcription in differentiating keratinocytes
Abstract
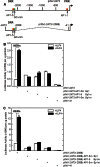
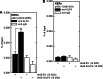
KLF4 is a member of the Kruppel-like factor family of transcriptional regulators. KLF4 has been shown to be required for normal terminal differentiation of keratinocytes, but the molecular mechanism whereby KLF4 regulates genes associated with the differentiation process has not been studied. In the present study, we explore the impact of KLF4 on expression of involucrin, a gene that is specifically expressed in differentiated keratinocytes. KLF4 overexpression and knockdown studies show that involucrin mRNA and protein level correlates directly with KLF4 level. Moreover, studies of mutant KLF4 proteins indicate that transcriptionally inactive forms do not increase involucrin expression. PKCδ is a regulator of keratinocyte differentiation that increases expression of differentiation-associated target genes, including involucrin. Overexpression of KLF4 augments the PKCδ-dependent increase in involucrin expression, whereas KLF4 knockdown attenuates this response. The KLF4 induction of human involucrin (hINV) promoter activity is mediated via KLF4 binding to a GC-rich element located in the hINV promoter distal regulatory region, a region of the promoter required for in vivo involucrin expression. Mutation of the GC-rich element, an adjacent AP1 factor binding site, or both sites severely attenuates the response. Moreover, loss of KLF4 in an epidermal equivalent model of differentiation results in loss of hINV expression. These studies suggest that KLF4 is part of a multiprotein complex that interacts that the hINV promoter distal regulatory region to drive differentiation-dependent hINV gene expression in epidermis.
Keywords: Cell Differentiation; Gene Expression; Keratinocytes; Kruppel-like Factor (KLF); Protein Kinase C (PKC).
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

