A powerful and efficient set test for genetic markers that handles confounders
- PMID: 23599503
- PMCID: PMC3673214
- DOI: 10.1093/bioinformatics/btt177
A powerful and efficient set test for genetic markers that handles confounders
Abstract
Motivation: Approaches for testing sets of variants, such as a set of rare or common variants within a gene or pathway, for association with complex traits are important. In particular, set tests allow for aggregation of weak signal within a set, can capture interplay among variants and reduce the burden of multiple hypothesis testing. Until now, these approaches did not address confounding by family relatedness and population structure, a problem that is becoming more important as larger datasets are used to increase power.
Results: We introduce a new approach for set tests that handles confounders. Our model is based on the linear mixed model and uses two random effects-one to capture the set association signal and one to capture confounders. We also introduce a computational speedup for two random-effects models that makes this approach feasible even for extremely large cohorts. Using this model with both the likelihood ratio test and score test, we find that the former yields more power while controlling type I error. Application of our approach to richly structured Genetic Analysis Workshop 14 data demonstrates that our method successfully corrects for population structure and family relatedness, whereas application of our method to a 15 000 individual Crohn's disease case-control cohort demonstrates that it additionally recovers genes not recoverable by univariate analysis.
Availability: A Python-based library implementing our approach is available at http://mscompbio.codeplex.com.
Figures
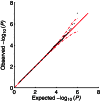
 ,1], where
,1], where  is the estimated mixing weight in the null distribution
is the estimated mixing weight in the null distributionSimilar articles
-
Greater power and computational efficiency for kernel-based association testing of sets of genetic variants.Bioinformatics. 2014 Nov 15;30(22):3206-14. doi: 10.1093/bioinformatics/btu504. Epub 2014 Jul 29. Bioinformatics. 2014. PMID: 25075117 Free PMC article.
-
Powerful Tests for Multi-Marker Association Analysis Using Ensemble Learning.PLoS One. 2015 Nov 30;10(11):e0143489. doi: 10.1371/journal.pone.0143489. eCollection 2015. PLoS One. 2015. PMID: 26619286 Free PMC article.
-
RL-SKAT: An Exact and Efficient Score Test for Heritability and Set Tests.Genetics. 2017 Dec;207(4):1275-1283. doi: 10.1534/genetics.117.300395. Epub 2017 Oct 12. Genetics. 2017. PMID: 29025915 Free PMC article.
-
Integrate multiple traits to detect novel trait-gene association using GWAS summary data with an adaptive test approach.Bioinformatics. 2019 Jul 1;35(13):2251-2257. doi: 10.1093/bioinformatics/bty961. Bioinformatics. 2019. PMID: 30476000 Free PMC article.
-
Population structure in genetic studies: Confounding factors and mixed models.PLoS Genet. 2018 Dec 27;14(12):e1007309. doi: 10.1371/journal.pgen.1007309. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30589851 Free PMC article. Review.
Cited by
-
The benefits of selecting phenotype-specific variants for applications of mixed models in genomics.Sci Rep. 2013;3:1815. doi: 10.1038/srep01815. Sci Rep. 2013. PMID: 23657357 Free PMC article.
-
networkGWAS: a network-based approach to discover genetic associations.Bioinformatics. 2023 Jun 1;39(6):btad370. doi: 10.1093/bioinformatics/btad370. Bioinformatics. 2023. PMID: 37285313 Free PMC article.
-
Quantifying missing heritability at known GWAS loci.PLoS Genet. 2013;9(12):e1003993. doi: 10.1371/journal.pgen.1003993. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385918 Free PMC article.
-
Robust Multi-Network Clustering via Joint Cross-Domain Cluster Alignment.Proc IEEE Int Conf Data Min. 2015 Nov;2015:291-300. doi: 10.1109/ICDM.2015.13. Proc IEEE Int Conf Data Min. 2015. PMID: 27239167 Free PMC article.
-
Using controls to limit false discovery in the era of big data.BMC Bioinformatics. 2018 Sep 14;19(1):323. doi: 10.1186/s12859-018-2356-2. BMC Bioinformatics. 2018. PMID: 30217148 Free PMC article.
References
-
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 2009;24:451–471.
-
- Balding DJ. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006;7:781–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

