Architecture and assembly of the Gram-positive cell wall
- PMID: 23600697
- PMCID: PMC3663049
- DOI: 10.1111/mmi.12203
Architecture and assembly of the Gram-positive cell wall
Abstract
The bacterial cell wall is a mesh polymer of peptidoglycan--linear glycan strands cross-linked by flexible peptides--that determines cell shape and provides physical protection. While the glycan strands in thin 'Gram-negative' peptidoglycan are known to run circumferentially around the cell, the architecture of the thicker 'Gram-positive' form remains unclear. Using electron cryotomography, here we show that Bacillus subtilis peptidoglycan is a uniformly dense layer with a textured surface. We further show it rips circumferentially, curls and thickens at free edges, and extends longitudinally when denatured. Molecular dynamics simulations show that only atomic models based on the circumferential topology recapitulate the observed curling and thickening, in support of an 'inside-to-outside' assembly process. We conclude that instead of being perpendicular to the cell surface or wrapped in coiled cables (two alternative models), the glycan strands in Gram-positive cell walls run circumferentially around the cell just as they do in Gram-negative cells. Together with providing insights into the architecture of the ultimate determinant of cell shape, this study is important because Gram-positive peptidoglycan is an antibiotic target crucial to the viability of several important rod-shaped pathogens including Bacillus anthracis, Listeria monocytogenes, and Clostridium difficile.
© 2013 John Wiley & Sons Ltd.
Figures



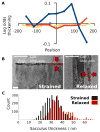
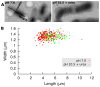
Comment in
-
Visualizing the production and arrangement of peptidoglycan in Gram-positive cells.Mol Microbiol. 2013 May;88(4):645-9. doi: 10.1111/mmi.12212. Epub 2013 Apr 1. Mol Microbiol. 2013. PMID: 23551458
References
-
- Chen Songye, McDowall Alasdair, Dobro Megan J, Briegel Ariane, Ladinsky Mark, Shi Jian, Tocheva Elitza I, et al. Electron Cryotomography of Bacterial Cells. Journal of Visualized Experiments: JoVE. 2010;(39) doi: 10.3791/1943. http://www.ncbi.nlm.nih.gov/pubmed/20461053. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

