Adult T-cell leukemia cells are characterized by abnormalities of Helios expression that promote T cell growth
- PMID: 23600753
- PMCID: PMC7657222
- DOI: 10.1111/cas.12181
Adult T-cell leukemia cells are characterized by abnormalities of Helios expression that promote T cell growth
Abstract
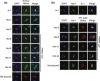
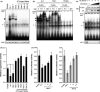
Molecular abnormalities involved in the multistep leukemogenesis of adult T-cell leukemia (ATL) remain to be clarified. Based on our integrated database, we focused on the expression patterns and levels of Ikaros family genes, Ikaros, Helios, and Aiolos, in ATL patients and HTLV-1 carriers. The results revealed profound deregulation of Helios expression, a pivotal regulator in the control of T-cell differentiation and activation. The majority of ATL samples (32/37 cases) showed abnormal splicing of Helios expression, and four cases did not express Helios. In addition, novel genomic loss in Helios locus was observed in 17/168 cases. We identified four ATL-specific short Helios isoforms and revealed their dominant-negative function. Ectopic expression of ATL-type Helios isoform as well as knockdown of normal Helios or Ikaros promoted T-cell growth. Global mRNA profiling and pathway analysis showed activation of several signaling pathways important for lymphocyte proliferation and survival. These data provide new insights into the molecular involvement of Helios function in the leukemogenesis and phenotype of ATL cells, indicating that Helios deregulation is one of the novel molecular hallmarks of ATL.
© 2013 Japanese Cancer Association.
Figures





References
-
- Yamaguchi K, Watanabe T. Human T lymphotropic virus type‐I and adult T‐cell leukemia in Japan. Int J Hematol 2002; 76: 240–45. - PubMed
-
- Iwanaga M, Watanabe T, Utsunomiya A et al Human T‐cell leukemia virus type I (HTLV‐1) proviral load and disease progression in asymptomatic HTLV‐1 carriers: a nationwide prospective study in Japan. Blood 2010; 116: 1211–19. - PubMed
-
- Yamagishi M, Nakano K, Miyake A et al Polycomb‐mediated loss of miR‐31 activates NIK‐dependent NF‐κB pathway in adult T cell leukemia and other cancers. Cancer Cell 2012; 21: 121–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

