Identification and biochemical characterization of Laodelphax striatellus neutral ceramidase
- PMID: 23601004
- PMCID: PMC3879266
- DOI: 10.1111/imb.12028
Identification and biochemical characterization of Laodelphax striatellus neutral ceramidase
Abstract
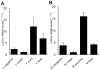
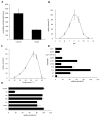
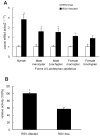
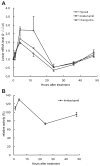
Ceramidases are a group of enzymes that catalyse hydrolysis of ceramides to generate fatty acid and sphingosine. In this study, we report the cloning and characterization of the rice small brown planthopper Laodelphax striatellus neutral ceramidase (nCDase), LsnCer. LsnCer was identified by sequencing the transcriptome of L. striatellus and is a protein of 717 amino acids with a predicted molecular weight of 79.3 kDa. Similarly to other known nCDases, the optimum pH for LsnCer is 8.0 and the optimum temperature is 37 °C for its in vitro activity. LsnCer activity is inhibited by Zn(2+) significantly and Fe(2+) slightly. LsnCer has broad substrate specificity with a preference for ceramides with a medium acyl-chain or a monounsaturated long acyl-chain. Infection with rice strip virus (RSV) or treatment with insecticides significantly increased LsnCer mRNA expression and its enzymatic activity in L. striatellus. These results suggest that LsnCer is a bona fide nCDase that may have a role in adaption of L. striatellus to environmental stresses.
Keywords: LsnCer; activity; insecticide; mRNA; nCDase; rice strip virus.
© 2013 Royal Entomological Society.
Figures



Similar articles
-
Rice Stripe Virus Infection Alters mRNA Levels of Sphingolipid-Metabolizing Enzymes and Sphingolipids Content in Laodelphax striatellus.J Insect Sci. 2017 Jan 27;17(1):16. doi: 10.1093/jisesa/iew111. Print 2017 Jan. J Insect Sci. 2017. PMID: 28130458 Free PMC article.
-
Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV).BMC Genomics. 2010 May 13;11:303. doi: 10.1186/1471-2164-11-303. BMC Genomics. 2010. PMID: 20462456 Free PMC article.
-
Ubiquitin-Conjugating Enzyme E2 E Inhibits the Accumulation of Rice Stripe Virus in Laodelphax striatellus (Fallén).Viruses. 2020 Aug 19;12(9):908. doi: 10.3390/v12090908. Viruses. 2020. PMID: 32825037 Free PMC article.
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
-
Role of Ceramidases in Sphingolipid Metabolism and Human Diseases.Cells. 2019 Dec 4;8(12):1573. doi: 10.3390/cells8121573. Cells. 2019. PMID: 31817238 Free PMC article. Review.
Cited by
-
A neutral ceramidase, NlnCDase, is involved in the stress responses of brown planthopper, Nilaparvata lugens (Stål).Sci Rep. 2018 Jan 18;8(1):1130. doi: 10.1038/s41598-018-19219-y. Sci Rep. 2018. PMID: 29348442 Free PMC article.
-
Neutral Ceramidase Is Required for the Reproduction of Brown Planthopper, Nilaparvata lugens (Stål).Front Physiol. 2021 Feb 24;12:629532. doi: 10.3389/fphys.2021.629532. eCollection 2021. Front Physiol. 2021. PMID: 33716775 Free PMC article.
-
Rice Stripe Virus Infection Alters mRNA Levels of Sphingolipid-Metabolizing Enzymes and Sphingolipids Content in Laodelphax striatellus.J Insect Sci. 2017 Jan 27;17(1):16. doi: 10.1093/jisesa/iew111. Print 2017 Jan. J Insect Sci. 2017. PMID: 28130458 Free PMC article.
-
Alkaline ceramidase (ClAC) inhibition enhances heat stress response in Cyrtorhinus lividipennis (Reuter).Front Physiol. 2023 May 9;14:1160846. doi: 10.3389/fphys.2023.1160846. eCollection 2023. Front Physiol. 2023. PMID: 37234408 Free PMC article.
-
Study of tyramine-binding mechanism and insecticidal activity of oil extracted from Eucalyptus against Sitophilus oryzae.Front Chem. 2022 Sep 23;10:964700. doi: 10.3389/fchem.2022.964700. eCollection 2022. Front Chem. 2022. PMID: 36212071 Free PMC article.
References
-
- Bao Y, Li B, Liu Z, Xue J, Zhu Z, Cheng J, Zhang C. Triazophos up-regulated gene expression in the female brown planthopper, Nilaparvata lugens. J Insect Physiol. 2010;56:1087–94. - PubMed
-
- Bar J, Linke T, Ferlinz K, Neumann U, Schuchman E, Sandhoff K. Molecular analysis of acid ceramidase deficiency in patients with Farber disease. Hum Mutat. 2001;17:199–209. - PubMed
-
- Choi M, Anderson M, Zhang Z, Zimonjic D, Popescu N, Mukherjee A. Neutral ceamidase gene: role in regulating ceramide-induced apoptosis. Gene. 2003;315:113–22. - PubMed
-
- El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters J, Hannun Y. Molecular cloning and characterization of a human mitochondrial ceramidase. J Biol Chem. 2000;275:21508–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

