Convergent evolution of [D-Leucine(1)] microcystin-LR in taxonomically disparate cyanobacteria
- PMID: 23601305
- PMCID: PMC3640908
- DOI: 10.1186/1471-2148-13-86
Convergent evolution of [D-Leucine(1)] microcystin-LR in taxonomically disparate cyanobacteria
Abstract
Background: Many important toxins and antibiotics are produced by non-ribosomal biosynthetic pathways. Microcystins are a chemically diverse family of potent peptide toxins and the end-products of a hybrid NRPS and PKS secondary metabolic pathway. They are produced by a variety of cyanobacteria and are responsible for the poisoning of humans as well as the deaths of wild and domestic animals around the world. The chemical diversity of the microcystin family is attributed to a number of genetic events that have resulted in the diversification of the pathway for microcystin assembly.
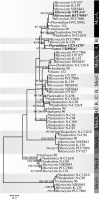
Results: Here, we show that independent evolutionary events affecting the substrate specificity of the microcystin biosynthetic pathway have resulted in convergence on a rare [D-Leu(1)] microcystin-LR chemical variant. We detected this rare microcystin variant from strains of the distantly related genera Microcystis, Nostoc, and Phormidium. Phylogenetic analysis performed using sequences of the catalytic domains within the mcy gene cluster demonstrated a clear recombination pattern in the adenylation domain phylogenetic tree. We found evidence for conversion of the gene encoding the McyA(2) adenylation domain in strains of the genera Nostoc and Phormidium. However, point mutations affecting the substrate-binding sequence motifs of the McyA(2) adenylation domain were associated with the change in substrate specificity in two strains of Microcystis. In addition to the main [D-Leu(1)] microcystin-LR variant, these two strains produced a new microcystin that was identified as [Met(1)] microcystin-LR.
Conclusions: Phylogenetic analysis demonstrated that both point mutations and gene conversion result in functional mcy gene clusters that produce the same rare [D-Leu(1)] variant of microcystin in strains of the genera Microcystis, Nostoc, and Phormidium. Engineering pathways to produce recombinant non-ribosomal peptides could provide new natural products or increase the activity of known compounds. Our results suggest that the replacement of entire adenylation domains could be a more successful strategy to obtain higher specificity in the modification of the non-ribosomal peptides than point mutations.
Figures






References
-
- Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–534. - PubMed
-
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. - PubMed
-
- Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. - PubMed
-
- Kallow W, Neuhof T, Arezi B, Jungblut P, Von Döhren H. Penicillin biosynthesis: intermediates of biosynthesis of delta-alpha-aminoadipyl-cysteinyl-valine formed by ACV synthetase from Acremonium chrysogenum. FEBS Lett. 1997;414:74–78. - PubMed
-
- Steenbergen J, Alder J, Thorne G, Tally F. Daptomycin: a lipopeptide antibiotic for the treatment of serious gram-positive infections. J Antimicrob Chemother. 2005;55:283–288. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

