Flux through trehalose synthase flows from trehalose to the alpha anomer of maltose in mycobacteria
- PMID: 23601637
- PMCID: PMC3918855
- DOI: 10.1016/j.chembiol.2013.02.014
Flux through trehalose synthase flows from trehalose to the alpha anomer of maltose in mycobacteria
Abstract
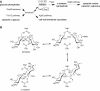
Trehalose synthase (TreS) was thought to catalyze flux from maltose to trehalose, a precursor of essential trehalose mycolates in mycobacterial cell walls. However, we now show, using a genetic approach, that TreS is not required for trehalose biosynthesis in Mycobacterium smegmatis, whereas two alternative trehalose-biosynthetic pathways (OtsAB and TreYZ) are crucial. Consistent with this direction of flux, trehalose levels in Mycobacterium tuberculosis decreased when TreS was overexpressed. In addition, TreS was shown to interconvert the α anomer of maltose and trehalose using (1)H and (19)F-nuclear magnetic resonance spectroscopies using its normal substrates and deoxyfluoromaltose analogs, with the nonenzymatic mutarotation of α/β-maltose being slow. Therefore, flux through TreS in mycobacteria flows from trehalose to α-maltose, which is the appropriate anomer for maltose kinase of the GlgE α-glucan pathway, which in turn contributes to intracellular and/or capsular polysaccharide biosynthesis.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




References
-
- Argüelles J.C. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 2000;174:217–224. - PubMed
-
- Bailey J.M., Fishman P.H., Pentchev P.G. Studies on mutarotases. I. Purification and properties of a mutarotase from higher plants. J. Biol. Chem. 1967;242:4263–4269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/000C0618/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000C0647/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F017294/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004561/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

