miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs
- PMID: 23601657
- PMCID: PMC3672661
- DOI: 10.1186/bcr3415
miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs
Abstract
Introduction: Breast cancer is a genetically and phenotypically complex disease. To understand the role of miRNAs in this molecular complexity, we performed miRNA expression analysis in a cohort of molecularly well-characterized human breast cancer cell lines to identify miRNAs associated with the most common molecular subtypes and the most frequent genetic aberrations.
Methods: Using a microarray carrying LNA™ modified oligonucleotide capture probes), expression levels of 725 human miRNAs were measured in 51 breast cancer cell lines. Differential miRNA expression was explored by unsupervised cluster analysis and was then associated with the molecular subtypes and genetic aberrations commonly present in breast cancer.
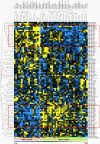
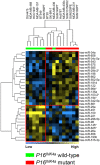
Results: Unsupervised cluster analysis using the most variably expressed miRNAs divided the 51 breast cancer cell lines into a major and a minor cluster predominantly mirroring the luminal and basal intrinsic subdivision of breast cancer cell lines. One hundred and thirteen miRNAs were differentially expressed between these two main clusters. Forty miRNAs were differentially expressed between basal-like and normal-like/claudin-low cell lines. Within the luminal-group, 39 miRNAs were associated with ERBB2 overexpression and 24 with E-cadherin gene mutations, which are frequent in this subtype of breast cancer cell lines. In contrast, 31 miRNAs were associated with E-cadherin promoter hypermethylation, which, contrary to E-cadherin mutation, is exclusively observed in breast cancer cell lines that are not of luminal origin. Thirty miRNAs were associated with p16INK4 status while only a few miRNAs were associated with BRCA1, PIK3CA/PTEN and TP53 mutation status. Twelve miRNAs were associated with DNA copy number variation of the respective locus.
Conclusion: Luminal-basal and epithelial-mesenchymal associated miRNAs determine the subdivision of miRNA transcriptome of breast cancer cell lines. Specific sets of miRNAs were associated with ERBB2 overexpression, p16INK4a or E-cadherin mutation or E-cadherin methylation status, which implies that these miRNAs may contribute to the driver role of these genetic aberrations. Additionally, miRNAs, which are located in a genomic region showing recurrent genetic aberrations, may themselves play a driver role in breast carcinogenesis or contribute to a driver gene in their vicinity. In short, our study provides detailed molecular miRNA portraits of breast cancer cell lines, which can be exploited for functional studies of clinically important miRNAs.
Figures




References
-
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. - PubMed
-
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, Mäkelä R, Naume B, Perälä M, Kallioniemi O, Kristensen VN, Yakhini Z, Børresen-Dale AL. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

