Sample preparation methodology for mouse heart metabolomics using comprehensive two-dimensional gas chromatography coupled with time-of-flight mass spectrometry
- PMID: 23601879
- PMCID: PMC3638982
- DOI: 10.1016/j.talanta.2013.03.005
Sample preparation methodology for mouse heart metabolomics using comprehensive two-dimensional gas chromatography coupled with time-of-flight mass spectrometry
Abstract
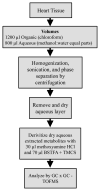
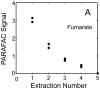
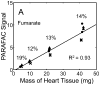
The investigation of naturally volatile and derivatized metabolites in mammalian tissues by comprehensive two-dimensional (2D) gas chromatography coupled with time-of-flight mass spectrometry (GC × GC-TOFMS) can provide the data for a comprehensive analysis of the pathophysiology of disease processes. When relative quantification is needed for hypothesis testing, the preparation of sample tissue must provide clear evidence that a quantitative relationship exists between the final detected signal and the amount of metabolite in the tissue. Herein, we report the optimization of a metabolite extraction method for mouse heart tissue for GC × GC-TOFMS analysis. A recursive extraction experiment was initially performed to measure the extraction efficiency of representative target metabolites (sugars, tricarboxylic acid cycle metabolites, amino acids, lipid and signaling molecules) in the aqueous fraction of a three-phase extraction system involving tissue, methanol:water, and chloroform. Some metabolites suffered from incomplete extraction with a single extraction of ≈ 40 mg in 600 μl organic and 400 μl aqueous phases, possibly caused by saturation effects. Subsequent experiments, calibrating resulting metabolite signal to the mass of heart tissue extracted, demonstrated that doubling the solvent volumes and a lower tissue mass was needed to provide accurate relative quantification of the derivatized mouse heart metabolome. We demonstrate quantitative extraction of metabolites from ≈ 20 mg of heart tissue using 1200 μl organic phase (chloroform) and 800 μl aqueous phase (methanol:water in equal parts by volume).
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures














Similar articles
-
The combination of four analytical methods to explore skeletal muscle metabolomics: Better coverage of metabolic pathways or a marketing argument?J Pharm Biomed Anal. 2018 Jan 30;148:273-279. doi: 10.1016/j.jpba.2017.10.013. Epub 2017 Oct 18. J Pharm Biomed Anal. 2018. PMID: 29059617
-
Solid phase microextraction coupled with comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry for high-resolution metabolite profiling in apples: implementation of structured separations for optimization of sample preparation procedure in complex samples.J Chromatogr A. 2012 Aug 17;1251:208-218. doi: 10.1016/j.chroma.2012.06.052. Epub 2012 Jun 23. J Chromatogr A. 2012. PMID: 22771060
-
A systematic evaluation of quenching and extraction procedures for quantitative metabolome profiling of HeLa carcinoma cell under 2D and 3D cell culture conditions.Biotechnol J. 2023 May;18(5):e2200444. doi: 10.1002/biot.202200444. Epub 2023 Feb 24. Biotechnol J. 2023. PMID: 36796787
-
Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols.Anal Bioanal Chem. 2011 Jan;399(3):1127-39. doi: 10.1007/s00216-010-4425-x. Epub 2010 Dec 2. Anal Bioanal Chem. 2011. PMID: 21125262
-
Methods of discovery-based and targeted metabolite analysis by comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry detection.Methods Mol Biol. 2014;1198:83-97. doi: 10.1007/978-1-4939-1258-2_6. Methods Mol Biol. 2014. PMID: 25270924 Review.
Cited by
-
Defective Branched-Chain Amino Acid Catabolism Disrupts Glucose Metabolism and Sensitizes the Heart to Ischemia-Reperfusion Injury.Cell Metab. 2017 Feb 7;25(2):374-385. doi: 10.1016/j.cmet.2016.11.005. Cell Metab. 2017. PMID: 28178567 Free PMC article.
-
Amino acid primed mTOR activity is essential for heart regeneration.iScience. 2021 Dec 6;25(1):103574. doi: 10.1016/j.isci.2021.103574. eCollection 2022 Jan 21. iScience. 2021. PMID: 34988408 Free PMC article.
-
Experimental Periodontitis Results in Prediabetes and Metabolic Alterations in Brain, Liver and Heart: Global Untargeted Metabolomic Analyses.J Oral Biol (Northborough). 2016;3(1):10.13188/2377-987X.1000020. doi: 10.13188/2377-987X.1000020. Epub 2016 Apr 23. J Oral Biol (Northborough). 2016. PMID: 27390783 Free PMC article.
-
Optimizing 2D gas chromatography mass spectrometry for robust tissue, serum and urine metabolite profiling.Talanta. 2017 Apr 1;165:685-691. doi: 10.1016/j.talanta.2017.01.003. Epub 2017 Jan 7. Talanta. 2017. PMID: 28153317 Free PMC article.
-
Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics.Trends Analyt Chem. 2018 Dec;109:275-286. doi: 10.1016/j.trac.2018.10.015. Epub 2018 Oct 24. Trends Analyt Chem. 2018. PMID: 30662103 Free PMC article.
References
-
- Rocha SM, Caldeira M, Carrola J, Santos M, Cruz N, Duarte IF. J Chromatogr A. 2012;1252:155–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

