The polymerase of negative-stranded RNA viruses
- PMID: 23602472
- PMCID: PMC4159711
- DOI: 10.1016/j.coviro.2013.03.008
The polymerase of negative-stranded RNA viruses
Abstract
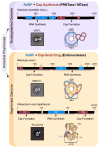
Negative-sense (NS) RNA viruses deliver into cells a mega-dalton RNA-protein complex competent for transcription. Within this complex, the RNA is protected in a nucleocapsid protein (NP) sheath which the viral polymerase negotiates during RNA synthesis. The NP-RNA templates come as nonsegmented (NNS) or segmented (SNS), necessitating distinct strategies for transcription by their polymerases. Atomic-level understanding of the NP-RNA of both NNS and SNS RNA viruses show that the RNA must be transiently dissociated from NP during RNA synthesis. Here we summarize and compare the polymerases of NNS and SNS RNA viruses, and the current structural data on the polymerases. Those comparisons inform us on the evolution of related RNA synthesis machines which use two distinct mechanisms for mRNA cap formation.
Copyright © 2013. Published by Elsevier B.V.
Figures



References
-
- Geisbert TW, Jahrling PB. Exotic emerging viral diseases: progress and challenges. Nat Med. 2004;10:S110–121. - PubMed
-
- Pappu HR, Jones RA, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141:219–236. - PubMed
-
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. - PubMed
-
- Morin B, Whelan SP. La protéine L des Mononegavirales. Virologie. 2012;16:258–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

