Signaling pathways that control mRNA turnover
- PMID: 23602935
- PMCID: PMC3703460
- DOI: 10.1016/j.cellsig.2013.03.026
Signaling pathways that control mRNA turnover
Abstract
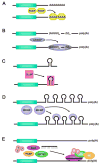
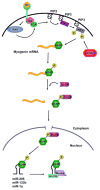
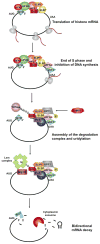
Cells regulate their genomes mainly at the level of transcription and at the level of mRNA decay. While regulation at the level of transcription is clearly important, the regulation of mRNA turnover by signaling networks is essential for a rapid response to external stimuli. Signaling pathways result in posttranslational modification of RNA binding proteins by phosphorylation, ubiquitination, methylation, acetylation etc. These modifications are important for rapid remodeling of dynamic ribonucleoprotein complexes and triggering mRNA decay. Understanding how these posttranslational modifications alter gene expression is therefore a fundamental question in biology. In this review we highlight recent findings on how signaling pathways and cell cycle checkpoints involving phosphorylation, ubiquitination, and arginine methylation affect mRNA turnover.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Garneau NL, Wilusz J, Wilusz CJ. Nat Rev Mol Cell Biol. 2007;8:113–126. - PubMed
-
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Nature. 2011;473:337–342. - PubMed
-
- Moore MJ, Proudfoot NJ. Cell. 2009;136:688–700. - PubMed
-
- Bellofatto V, Wilusz J. Cell. 2011;147:1438–1439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

