Ribonucleotides are signals for mismatch repair of leading-strand replication errors
- PMID: 23603118
- PMCID: PMC3658170
- DOI: 10.1016/j.molcel.2013.03.017
Ribonucleotides are signals for mismatch repair of leading-strand replication errors
Abstract
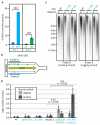
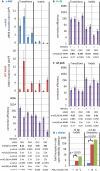
To maintain genome stability, mismatch repair of nuclear DNA replication errors must be directed to the nascent strand, likely by DNA ends and PCNA. Here we show that the efficiency of mismatch repair in Saccharomyces cerevisiae is reduced by inactivating RNase H2, which nicks DNA containing ribonucleotides incorporated during replication. In strains encoding mutator polymerases, this reduction is preferential for repair of mismatches made by leading-strand DNA polymerase ε as compared to lagging-strand DNA polymerase δ. The results suggest that RNase-H2-dependent processing of ribonucleotides transiently present in DNA after replication may direct mismatch repair to the continuously replicated nascent leading strand.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
-
- Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19756–19762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

