The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression
- PMID: 23603901
- PMCID: PMC3656328
- DOI: 10.1101/gad.217778.113
The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression
Abstract
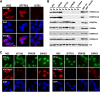
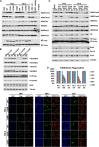
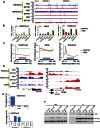
Recent studies have identified a Lys 27-to-methionine (K27M) mutation at one allele of H3F3A, one of the two genes encoding histone H3 variant H3.3, in 60% of high-grade pediatric glioma cases. The median survival of this group of patients after diagnosis is ∼1 yr. Here we show that the levels of H3K27 di- and trimethylation (H3K27me2 and H3K27me3) are reduced globally in H3.3K27M patient samples due to the expression of the H3.3K27M mutant allele. Remarkably, we also observed that H3K27me3 and Ezh2 (the catalytic subunit of H3K27 methyltransferase) at chromatin are dramatically increased locally at hundreds of gene loci in H3.3K27M patient cells. Moreover, the gain of H3K27me3 and Ezh2 at gene promoters alters the expression of genes that are associated with various cancer pathways. These results indicate that H3.3K27M mutation reprograms epigenetic landscape and gene expression, which may drive tumorigenesis.
Keywords: H3.3K27M; H3K27 methylation; PRC2; gliomas; pediatric.
Figures

References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Buckner JC 2003. Factors influencing survival in high-grade gliomas. Semin Oncol 30: 10–14 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
