Species-specific detection of the antiviral small-molecule compound CMA by STING
- PMID: 23604073
- PMCID: PMC3655471
- DOI: 10.1038/emboj.2013.86
Species-specific detection of the antiviral small-molecule compound CMA by STING
Abstract
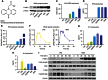
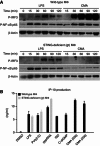
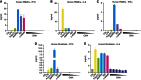
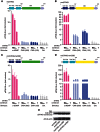
Extensive research on antiviral small molecules starting in the early 1970s has led to the identification of 10-carboxymethyl-9-acridanone (CMA) as a potent type I interferon (IFN) inducer. Up to date, the mode of action of this antiviral molecule has remained elusive. Here we demonstrate that CMA mediates a cell-intrinsic type I IFN response, depending on the ER-resident protein STING. CMA directly binds to STING and triggers a strong antiviral response through the TBK1/IRF3 route. Interestingly, while CMA displays extraordinary activity in phosphorylating IRF3 in the murine system, CMA fails to activate human cells that are otherwise responsive to STING ligands. This failure to activate human STING can be ascribed to its inability to bind to the C-terminal ligand-binding domain of human STING. Crystallographic studies show that two CMA molecules bind to the central Cyclic diguanylate (c-diGMP)-binding pocket of the STING dimer and fold the lid region in a fashion similar, but partially distinct, to c-diGMP. Altogether, these results provide novel insight into ligand-sensing properties of STING and, furthermore, unravel unexpected species-specific differences of this innate sensor.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
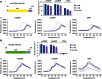
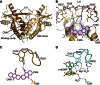
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 - PMC - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

