Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti
- PMID: 23604077
- PMCID: PMC3689809
- DOI: 10.1534/g3.113.006338
Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti
Abstract
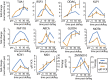
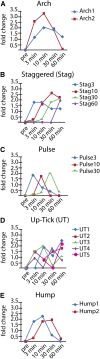
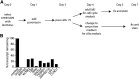
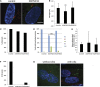
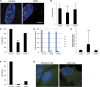
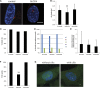
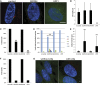
Cilia are microtubule based organelles that project from cells. Cilia are found on almost every cell type of the human body and numerous diseases, collectively termed ciliopathies, are associated with defects in cilia, including respiratory infections, male infertility, situs inversus, polycystic kidney disease, retinal degeneration, and Bardet-Biedl Syndrome. Here we show that Illumina-based whole-genome transcriptome analysis in the biflagellate green alga Chlamydomonas reinhardtii identifies 1850 genes up-regulated during ciliogenesis, 4392 genes down-regulated, and 4548 genes with no change in expression during ciliogenesis. We examined four genes up-regulated and not previously known to be involved with cilia (ZMYND10, NXN, GLOD4, SPATA4) by knockdown of the human orthologs in human retinal pigment epithelial cells (hTERT-RPE1) cells to ask whether they are involved in cilia-related processes that include cilia assembly, cilia length control, basal body/centriole numbers, and the distance between basal bodies/centrioles. All of the genes have cilia-related phenotypes and, surprisingly, our data show that knockdown of GLOD4 and SPATA4 also affects the cell cycle. These results demonstrate that whole-genome transcriptome analysis during ciliogenesis is a powerful tool to gain insight into the molecular mechanism by which centrosomes and cilia are assembled.
Keywords: GLOD4; NXN; SPATA4; ZMYND10; deflagellation; flagella.
Figures
References
-
- Albee A. J., Dutcher S. K., 2012. Cilia and Human Disease in eLS. John Wiley & Sons, Ltd, New York
-
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., et al. , 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117: 527–539 - PubMed
-
- Azzalin C. M., Lingner J., 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16: 433–439 - PubMed
-
- Brady S. M., Orlando D. A., Lee J. Y., Wang J. Y., Koch J., et al. , 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
