HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin
- PMID: 23604080
- PMCID: PMC3661211
- DOI: 10.1038/nsmb.2565
HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin
Abstract
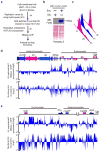
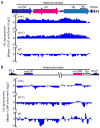
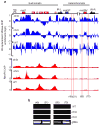
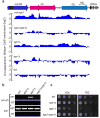
Heterochromatin causes epigenetic repression that can be transmitted through multiple cell divisions. However, the mechanisms underlying silencing and stability of heterochromatin are not fully understood. We show that heterochromatin differs from euchromatin in histone turnover and identify histone deacetylase (HDAC) Clr3 as a factor required for inhibiting histone turnover across heterochromatin domains in Schizosaccharomyces pombe. Loss of RNA-interference factors, Clr4 methyltransferase or HP1 proteins involved in HDAC localization causes increased histone turnover across pericentromeric domains. Clr3 also affects histone turnover at the silent mating-type region, where it can be recruited by alternative mechanisms acting in parallel to H3K9me-HP1. Notably, the JmjC-domain protein Epe1 promotes histone exchange, and loss of Epe1 suppresses both histone turnover and defects in heterochromatic silencing. Our results suggest that heterochromatic-silencing factors preclude histone turnover to promote silencing and inheritance of repressive chromatin.
Figures



References
-
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. - PubMed
-
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. - PubMed
-
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

