Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription
- PMID: 23604098
- PMCID: PMC3738159
- DOI: 10.1093/dnares/dst012
Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription
Abstract
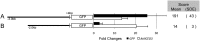
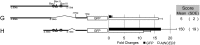
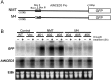
Plants respond to dehydration stress and tolerate water-deficit status through complex physiological and cellular processes. Many genes are induced by water deficit. Abscisic acid (ABA) plays important roles in tolerance to dehydration stress by inducing many stress genes. ABA is synthesized de novo in response to dehydration. Most of the genes involved in ABA biosynthesis have been identified, and they are expressed mainly in leaf vascular tissues. Of the products of such genes, 9-cis-epoxycarotenoid dioxygenase (NCED) is a key enzyme in ABA biosynthesis. One of the five NCED genes in Arabidopsis, AtNCED3, is significantly induced by dehydration. To understand the regulatory mechanism of the early stages of the dehydration stress response, it is important to analyse the transcriptional regulatory systems of AtNCED3. In the present study, we found that an overlapping G-box recognition sequence (5'-CACGTG-3') at -2248 bp from the transcriptional start site of AtNCED3 is an important cis-acting element in the induction of the dehydration response. We discuss the possible transcriptional regulatory system of dehydration-responsive AtNCED3 expression, and how this may control the level of ABA under water-deficit conditions.
Keywords: 9-cis-epoxycarotenoid dioxygenase (NCED); Arabidopsis; AtNCED3; abscisic acid (ABA); cis-acting element; dehydration response.
Figures




References
-
- Barnabas B., Jager K., Feher A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008;31:11–38. - PubMed
-
- Zhang J.Z., Creelman R.A., Zhu J.K. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 2004;135:615–21. doi:10.1104/pp.104.040295. - DOI - PMC - PubMed
-
- Reynolds M., Tuberosa R. Translational research impacting on crop productivity in drought-prone environments. Curr. Opin. Plant Biol. 2008;11:171–9. doi:10.1016/j.pbi.2008.02.005. - DOI - PubMed
-
- Moore J.P., Le N.T., Brandt W.F., Driouich A., Farrant J.M. Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 2009;14:110–7. doi:10.1016/j.tplants.2008.11.007. - DOI - PubMed
-
- Roy S.J., Tucker E.J., Tester M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011;14:232–9. doi:10.1016/j.pbi.2011.03.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

