STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells
- PMID: 23604114
- PMCID: PMC3868635
- DOI: 10.1038/onc.2013.115
STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells
Abstract
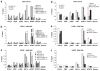
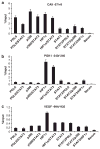
Solid tumors often exhibit simultaneously inflammatory and hypoxic microenvironments. The 'signal transducer and activator of transcription-3' (STAT3)-mediated inflammatory response and the hypoxia-inducible factor (HIF)-mediated hypoxia response have been independently shown to promote tumorigenesis through the activation of HIF or STAT3 target genes and to be indicative of a poor prognosis in a variety of tumors. We report here for the first time that STAT3 is involved in the HIF1, but not HIF2-mediated hypoxic transcriptional response. We show that inhibiting STAT3 activity in MDA-MB-231 and RCC4 cells by a STAT3 inhibitor or STAT3 small interfering RNA significantly reduces the levels of HIF1, but not HIF2 target genes in spite of normal levels of hypoxia-inducible transcription factor 1α (HIF1α) and HIF2α protein. Mechanistically, STAT3 activates HIF1 target genes by binding to HIF1 target gene promoters, interacting with HIF1α protein and recruiting coactivators CREB binding protein (CBP) and p300, and RNA polymerase II (Pol II) to form enhanceosome complexes that contain HIF1α, STAT3, CBP, p300 and RNA Pol II on HIF1 target gene promoters. Functionally, the effect of STAT3 knockdown on proliferation, motility and clonogenic survival of tumor cells in vitro is phenocopied by HIF1α knockdown in hypoxic cells, whereas STAT3 knockdown in normoxic cells also reduces cell proliferation, motility and clonogenic survival. This indicates that STAT3 works with HIF1 to activate HIF1 target genes and to drive HIF1-depedent tumorigenesis under hypoxic conditions, but also has HIF-independent activity in normoxic and hypoxic cells. Identifying the role of STAT3 in the hypoxia response provides further data supporting the effectiveness of STAT3 inhibitors in solid tumor treatment owing to their usefulness in inhibiting both the STAT3 and HIF1 pro-tumorigenic signaling pathways in some cancer types.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



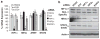


Similar articles
-
STAT3 or USF2 contributes to HIF target gene specificity.PLoS One. 2013 Aug 21;8(8):e72358. doi: 10.1371/journal.pone.0072358. eCollection 2013. PLoS One. 2013. PMID: 23991099 Free PMC article.
-
Upstream stimulatory factor 2 and hypoxia-inducible factor 2α (HIF2α) cooperatively activate HIF2 target genes during hypoxia.Mol Cell Biol. 2012 Nov;32(22):4595-610. doi: 10.1128/MCB.00724-12. Epub 2012 Sep 10. Mol Cell Biol. 2012. PMID: 22966206 Free PMC article.
-
BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes.Mol Cell Biol. 2013 Oct;33(19):3849-63. doi: 10.1128/MCB.00731-13. Epub 2013 Jul 29. Mol Cell Biol. 2013. PMID: 23897427 Free PMC article.
-
Enhanceosomes as integrators of hypoxia inducible factor (HIF) and other transcription factors in the hypoxic transcriptional response.Cell Signal. 2013 Sep;25(9):1895-903. doi: 10.1016/j.cellsig.2013.05.018. Epub 2013 May 21. Cell Signal. 2013. PMID: 23707522 Free PMC article. Review.
-
The role of HIF1α in renal cell carcinoma tumorigenesis.J Mol Med (Berl). 2014 Aug;92(8):825-36. doi: 10.1007/s00109-014-1180-z. Epub 2014 Jun 12. J Mol Med (Berl). 2014. PMID: 24916472 Free PMC article. Review.
Cited by
-
Under-expression of CK2β subunit in ccRCC represents a complementary biomarker of p-STAT3 Ser727 that correlates with patient survival.Oncotarget. 2017 Dec 19;9(5):5736-5751. doi: 10.18632/oncotarget.23422. eCollection 2018 Jan 19. Oncotarget. 2017. PMID: 29464030 Free PMC article.
-
Comprehensive pan-cancer analysis of STAT3 as a prognostic and immunological biomarker.Sci Rep. 2023 Mar 28;13(1):5069. doi: 10.1038/s41598-023-31226-2. Sci Rep. 2023. PMID: 36977736 Free PMC article.
-
Bile Acid Metabolism Mediates Cholesterol Homeostasis and Promotes Tumorigenesis in Clear Cell Renal Cell Carcinoma.Cancer Res. 2024 May 15;84(10):1570-1582. doi: 10.1158/0008-5472.CAN-23-0821. Cancer Res. 2024. PMID: 38417134 Free PMC article.
-
The key regulator circPDE3B promotes arsenic-induced bladder carcinogenesis by affecting STAT3 and NF-κB stability.Cell Biol Toxicol. 2025 May 28;41(1):91. doi: 10.1007/s10565-025-10038-2. Cell Biol Toxicol. 2025. PMID: 40437145 Free PMC article.
-
STAT3-Mediated Metabolic Reprograming in Cellular Transformation and Implications for Drug Resistance.Front Oncol. 2015 Jun 8;5:121. doi: 10.3389/fonc.2015.00121. eCollection 2015. Front Oncol. 2015. PMID: 26106584 Free PMC article. Review.
References
-
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

