RRP1B is a metastasis modifier that regulates the expression of alternative mRNA isoforms through interactions with SRSF1
- PMID: 23604122
- PMCID: PMC3925194
- DOI: 10.1038/onc.2013.133
RRP1B is a metastasis modifier that regulates the expression of alternative mRNA isoforms through interactions with SRSF1
Abstract
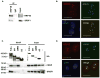
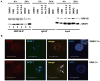
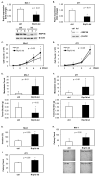
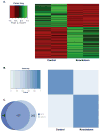
RRP1B (ribosomal RNA processing 1 homolog B) was first identified as a metastasis susceptibility gene in breast cancer through its ability to modulate gene expression in a manner that can be used to accurately predict prognosis in breast cancer. However, the mechanism(s) by which RRP1B modulates gene expression is currently unclear. Many RRP1B binding candidates are involved in alternative splicing, a mechanism of gene expression regulation that is increasingly recognized to be involved in cancer progression and metastasis. One such target is SRSF1 (serine/arginine-rich splicing factor 1) (SF2/ASF, splicing factor 2/alternative splicing factor), an essential splicing regulator that also functions as an oncoprotein. Earlier studies demonstrated that splicing and transcription occur concurrently and are coupled processes. Given that RRP1B regulates transcriptional activity, we hypothesized that RRP1B also regulates the expression of alternative mRNA isoforms through its interaction with SRSF1. Interaction between RRP1B and SRSF1 was verified by coimmunoprecipitation and coimmunofluorescence. Treatment of cells with transcriptional inhibitors significantly increased this interaction, demonstrating that the association of these two proteins is transcriptionally regulated. To assess the role of RRP1B in the regulation of alternative isoform expression, RNA-sequencing data were generated from control and Rrp1b-knockdown cells. Knockdown of Rrp1b induced a significant change in isoform expression in over 600 genes compared with control cell lines. This was verified by quantitative reverse-transcription PCR using isoform-specific primers. Pathway enrichment analyses identified cell cycle and checkpoint regulation to be those most affected by Rrp1b knockdown. These data suggest that RRP1B suppresses metastatic progression by altering the transcriptome through its interaction with splicing regulators such as SRSF1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003 Jan;33(1):49–54. - PubMed
-
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002 Dec 19;347(25):1999–2009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

