Heterogeneity of tumor-induced gene expression changes in the human metabolic network
- PMID: 23604282
- PMCID: PMC3681899
- DOI: 10.1038/nbt.2530
Heterogeneity of tumor-induced gene expression changes in the human metabolic network
Abstract
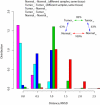
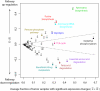
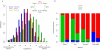
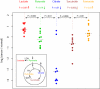
Reprogramming of cellular metabolism is an emerging hallmark of neoplastic transformation. However, it is not known how the expression of metabolic genes in tumors differs from that in normal tissues, or whether different tumor types exhibit similar metabolic changes. Here we compare expression patterns of metabolic genes across 22 diverse types of human tumors. Overall, the metabolic gene expression program in tumors is similar to that in the corresponding normal tissues. Although expression changes of some metabolic pathways (e.g., upregulation of nucleotide biosynthesis and glycolysis) are frequently observed across tumors, expression changes of other pathways (e.g., oxidative phosphorylation) are very heterogeneous. Our analysis also suggests that the expression changes of some metabolic genes (e.g., isocitrate dehydrogenase and fumarate hydratase) may enhance or mimic the effects of recurrent mutations in tumors. On the level of individual biochemical reactions, many hundreds of metabolic isoenzymes show significant and tumor-specific expression changes. These isoenzymes are potential targets for anticancer therapy.
Figures

Comment in
-
Cancer metabolism in breadth and depth.Nat Biotechnol. 2013 Jun;31(6):505-7. doi: 10.1038/nbt.2611. Nat Biotechnol. 2013. PMID: 23752435 No abstract available.
References
-
- Warburg O, Posener K, Negelein E. On the metabolism of carcinoma cells. Biochem Z. 1924;152:309–344.
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

