FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade
- PMID: 23604317
- PMCID: PMC3865866
- DOI: 10.1038/ncb2731
FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade
Abstract
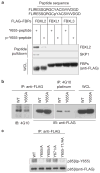
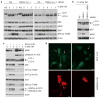
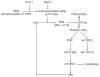
F-box proteins are the substrate-recognition subunits of SCF (Skp1/Cul1/F-box protein) ubiquitin ligase complexes. Purification of the F-box protein FBXL2 identified the PI(3)K regulatory subunit p85β and tyrosine phosphatase PTPL1 as interacting proteins. FBXL2 interacts with the pool of p85β that is free of p110 PI(3)K catalytic subunits and targets this pool for ubiquitylation and subsequent proteasomal degradation. FBXL2-mediated degradation of p85β is dependent on the integrity of its CaaX motif. Whereas most SCF substrates require phosphorylation to interact with their F-box proteins, phosphorylation of p85β on Tyr 655, which is adjacent to the degron, inhibits p85β binding to FBXL2. Dephosphorylation of phospho-Tyr-655 by PTPL1 stimulates p85β binding to and degradation through FBXL2. Finally, defects in the FBXL2-mediated degradation of p85β inhibit the binding of p110 subunits to IRS1, attenuate the PI(3)K signalling cascade and promote autophagy. We propose that FBXL2 and PTPL1 suppress p85β levels, preventing the inhibition of PI(3)K by an excess of free p85 that could compete with p85-p110 heterodimers for IRS1.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. - PubMed
-
- Skaar JR, Pagan JK, Pagano M. SnapShot: F box proteins I. Cell. 2009;137:1160–1161. - PubMed
-
- Skaar JR, D’Angiolella V, Pagan JK, Pagano M, SnapShot F. Box Proteins II. Cell. 2009;137:1358. - PubMed
-
- Wang C, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

