Fatty acids regulate perilipin5 in muscle by activating PPARδ
- PMID: 23606724
- PMCID: PMC3679396
- DOI: 10.1194/jlr.M038992
Fatty acids regulate perilipin5 in muscle by activating PPARδ
Abstract
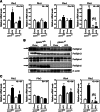
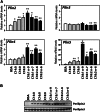
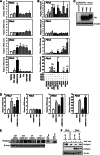
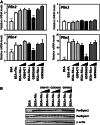
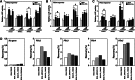
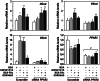
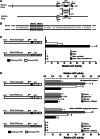
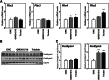
The surface of lipid droplets (LDs) in various cell types is coated with perilipin proteins encoded by the Plin genes. Perilipins regulate LD metabolism by selectively recruiting lipases and other proteins to LDs. We have studied the expression of perilipins in mouse muscle. The glycolytic fiber-enriched gastrocnemius muscle expresses predominantly Plin2-4. The oxidative fiber-enriched soleus muscle expresses Plin2-5. Expression of Plin2 and Plin4-5 is elevated in gastrocnemius and soleus muscles from mice fed a high-fat diet. This effect is preserved in peroxisome proliferator-activated receptor (PPAR)α-deficient mice. Mouse muscle derived C2C12 cells differentiated into glycolytic fibers increase transcription of these Plins when exposed to various long chain fatty acids (FAs). To understand how FAs regulate Plin genes, we used specific activators and antagonists against PPARs, Plin promoter reporter assays, chromatin immunoprecipitation, siRNA, and animal models. Our analyses demonstrate that FAs require PPARδ to induce transcription of Plin4 and Plin5. We further identify a functional PPAR binding site in the Plin5 gene and establish Plin5 as a novel direct PPARδ target in muscle. Our study reveals that muscle cells respond to elevated FAs by increasing transcription of several perilipin LD-coating proteins. This induction renders the muscle better equipped to sequester incoming FAs into cytosolic LDs.
Keywords: Lsdp5; Plin; Plin2; Plin3; Plin4; Plin5; S3-12; TIP47; oleic acid; perilipin; peroxisome proliferator-activated receptor response element; peroxisome proliferator-activated receptor δ; promoter.
Figures
References
-
- Eckardt K., Taube A., Eckel J. 2011. Obesity-associated insulin resistance in skeletal muscle: role of lipid accumulation and physical inactivity. Rev. Endocr. Metab. Disord. 12: 163–172 - PubMed
-
- Amati F., Dube J. J., Alvarez-Carnero E., Edreira M. M., Chomentowski P., Coen P. M., Switzer G. E., Bickel P. E., Stefanovic-Racic M., Toledo F. G., et al. 2011. Skeletal muscle triglycerides, diacylglycerols, and ceramides in insulin resistance: another paradox in endurance-trained athletes? Diabetes. 60: 2588–2597 - PMC - PubMed
-
- Brasaemle D. L. 2007. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

