Identifying residues that cause pH-dependent reduction potentials
- PMID: 23607577
- PMCID: PMC3691860
- DOI: 10.1021/bi4002858
Identifying residues that cause pH-dependent reduction potentials
Abstract
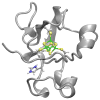
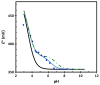
The pH dependence of the reduction potential E° for a metalloprotein indicates that the protonation state of at least one residue near the redox site changes and may be important for its activity. The responsible residue is usually identified by site-specific mutagenesis, which may be time-consuming. Here, the titration of E° for Chromatium vinosum high-potential iron-sulfur protein is predicted to be in good agreement with experiment using density functional theory and Poisson-Boltzmann calculations if only the sole histidine undergoes changes in protonation. The implementation of this approach into CHARMMing, a user-friendly web-based portal, allows users to identify residues in other proteins causing similar pH dependence.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
The role of histidine-42 in the oxidation-reduction mechanism of Chromatium vinosum high potential iron-sulfur protein.Biochim Biophys Acta. 1980 Dec 3;593(2):371-83. doi: 10.1016/0005-2728(80)90074-2. Biochim Biophys Acta. 1980. PMID: 7236640
-
The redox properties of the iron-sulphur cluster in hydrogenase from Chromatium vinosum, strain D.Biochimie. 1986 Jan;68(1):93-6. doi: 10.1016/s0300-9084(86)81073-2. Biochimie. 1986. PMID: 3015252
-
Dependence of the rates of dissolution of the Fe4S4 clusters of Chromatium vinosum high-potential iron protein and ferredoxin on cluster oxidation state.Proc Natl Acad Sci U S A. 1977 Dec;74(12):5231-4. doi: 10.1073/pnas.74.12.5231. Proc Natl Acad Sci U S A. 1977. PMID: 23530 Free PMC article.
-
Distinct redox behaviour of prosthetic groups in ready and unready hydrogenase from Chromatium vinosum.Biochim Biophys Acta. 1992 Feb 26;1119(2):157-68. doi: 10.1016/0167-4838(92)90386-r. Biochim Biophys Acta. 1992. PMID: 1540647
-
Coupling of electron transfer to proton uptake at the Q(B) site of the bacterial reaction center: a perspective from FTIR difference spectroscopy.Biochim Biophys Acta. 2008 Oct;1777(10):1229-48. doi: 10.1016/j.bbabio.2008.06.012. Epub 2008 Jul 11. Biochim Biophys Acta. 2008. PMID: 18671937 Review.
Cited by
-
Web-based computational chemistry education with CHARMMing III: Reduction potentials of electron transfer proteins.PLoS Comput Biol. 2014 Jul 24;10(7):e1003739. doi: 10.1371/journal.pcbi.1003739. eCollection 2014 Jul. PLoS Comput Biol. 2014. PMID: 25058418 Free PMC article.
-
Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers.Chem Rev. 2014 Apr 23;114(8):4366-469. doi: 10.1021/cr400479b. Chem Rev. 2014. PMID: 24758379 Free PMC article. Review. No abstract available.
-
Protein dynamics and the all-ferrous [Fe4 S4 ] cluster in the nitrogenase iron protein.Protein Sci. 2016 Jan;25(1):12-8. doi: 10.1002/pro.2772. Epub 2015 Sep 1. Protein Sci. 2016. PMID: 26271353 Free PMC article.
References
-
- Yang AS, Gunner MR, Sampogna R, Sharp K, Honig B. Proteins: Struct, Funct, Genet. 1993;15:252–265. - PubMed
-
- Russell ST, Warshel A. J Mol Biol. 1985;185:389–404. - PubMed
-
- Warshel A, Sussman F, King G. Biochemistry. 1986;25:8368–8372. - PubMed
-
- Bashford D, Karplus M. Biochemistry. 1990;29:10219–10225. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

