Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors
- PMID: 23607900
- PMCID: PMC3640902
- DOI: 10.1186/1471-2164-14-270
Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors
Abstract
Background: Wheat yellow (stripe) rust caused by Puccinia striiformis f. sp. tritici (PST) is one of the most devastating diseases of wheat worldwide. To design effective breeding strategies that maximize the potential for durable disease resistance it is important to understand the molecular basis of PST pathogenicity. In particular, the characterisation of the structure, function and evolutionary dynamics of secreted effector proteins that are detected by host immune receptors can help guide and prioritize breeding efforts. However, to date, our knowledge of the effector repertoire of cereal rust pathogens is limited.
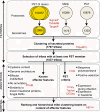
Results: We re-sequenced genomes of four PST isolates from the US and UK to identify effector candidates and relate them to their distinct virulence profiles. First, we assessed SNP frequencies between all isolates, with heterokaryotic SNPs being over tenfold more frequent (5.29 ± 2.23 SNPs/kb) than homokaryotic SNPs (0.41 ± 0.28 SNPs/kb). Next, we implemented a bioinformatics pipeline to integrate genomics, transcriptomics, and effector-focused annotations to identify and classify effector candidates in PST. RNAseq analysis highlighted transcripts encoding secreted proteins that were significantly enriched in haustoria compared to infected tissue. The expression of 22 candidate effector genes was characterised using qRT-PCR, revealing distinct temporal expression patterns during infection in wheat. Lastly, we identified proteins that displayed non-synonymous substitutions specifically between the two UK isolates PST-87/7 and PST-08/21, which differ in virulence to two wheat varieties. By focusing on polymorphic variants enriched in haustoria, we identified five polymorphic effector candidates between PST-87/7 and PST-08/21 among 2,999 secreted proteins. These allelic variants are now a priority for functional validation as virulence/avirulence effectors in the corresponding wheat varieties.
Conclusions: Integration of genomics, transcriptomics, and effector-directed annotation of PST isolates has enabled us to move beyond the single isolate-directed catalogues of effector proteins and develop a framework for mining effector proteins in closely related isolates and relate these back to their defined virulence profiles. This should ultimately lead to more comprehensive understanding of the PST pathogenesis system, an important first step towards developing more effective surveillance and management strategies for one of the most devastating pathogens of wheat.
Figures

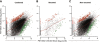

References
-
- Chen X. Epidemiology and control of stripe rust Puccinia striiformis f. sp. tritici on wheat. Canadian Journal of Plant Pathology. 2005;27(3):314–337. doi: 10.1080/07060660509507230. - DOI
-
- Wellings CR. Global status of stripe rust: a review of historical and current threats. Euphytica. 2011;179(1):129–141. doi: 10.1007/s10681-011-0360-y. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

